ABSTRACT
Leaves of Chionanthus retusus were found to be damaged by leaf spot disease associated with a fungus in Iksan, Korea. Leaf spots were angular to irregular, vein-limited, scattered, 1-8 mm diameter, brownish-gray to dark brown when dry, with heavy fructification. The pathogen causes premature defoliation of C. retusus plant and was identified as Pseudocercospora chionanthi-retusi based on morphological and molecular-phylogenetic analyses. The phylogenetic tree was constructed using multi-locus DNA sequence data of partial actin (actA), partial translation elongation factor 1-alfa (tef1), partial DNA-directed RNA polymerase II second largest subunit (rpb2) genes, and internal transcribed spacer regions. Current study provides detail morphological description of P. chionanthi-retusi on C. retusus in Korea, with supports of phylogenetic analysis and pathogenicity test.
Introduction
Chionanthus retusus Lindl. & Paxton (Oleaceae), known as a Chinese fringetree is a deciduous plant, natively grown in East Asia, particularly China, Japan, Korea, and Taiwan. The tree is planted for decorative purposes due to its white, rice-shaped flowers. From 2007 to 2013, during our routine field forays, leaves of
C. retusus were found to be damaged by leaf spot disease in Iksan, Korea. Further observations in several locations of the country revealed that leaf spot symptoms are associated with a fungus. Tracing of the literature showed that hitherto
C. retusus was recorded as host for several foliar fungal pathogens, such as
Aecidium fraxini-bungeanae Dietel,
Didymella macrostoma (Mont.) Qian Chen & L. Cai,
Discula fraxinea (Peck) Redlin & Stack.,
Erysiphe chionanthi (R. Y. Zheng & G. Q. Chen) U. Braun & S. Takam.,
Passalora chionanthi (Ellis & Everh.) U. Braun, and
Pseudocercospora chionanthi-retusi Goh & W. H. Hsieh. in China, Japan, Taiwan, Korea, and the United States (
Farr and Rossman, 2022). The purpose of the present study was to describe this fungus causing leaf spot disease of
C. retusus based on the morphological characteristics with an application of molecular-phylogenetic analysis using multilocus gene sequences and pathogenicity test.
Materials and Methods
Sample source. Totally, the following nine voucher specimens kept at the Korea University Herbarium were involved in this study: KUS-F231140 (Osan: 30 Oct 2007), F27686 (Iksan: 5 Oct 2013), F28182 (Iksan: 20 Sep 2014), F28206 (Jinju: 22 Sep 2014), F28207 (Jinju: 23 Sep 2014), F28256 (Busan: 25 Sep 2014), F28335 (Osan: 8 Oct 2014), F28991 (Gwangju: 23 Oct 2015), F30108 (Mokpo: 2 Oct 2017).
Morphological examination. Small cuts taken from lesions of infected leaves from KUS- F28182 and F27686 were mounted in a few drops of distilled water, then examined under an optic microscope (Olympus BX51, Tokyo, Japan, and Carl Zeiss AX10, Göttingen, Germany equipped with a KCS-3.1C imaging system) using bright-field and differential interference contrast. At least 50 measurements were taken for each structure.
Culture isolation. To obtain a single conidial isolate, the conidiophore was singly detached with a sterile needle from infected leaves of samples KUS-F27686, F28182, and F28206 and carefully placed onto 2% water agar (Junsei) amended with 100 mg/l of streptomycin sulfate. After 3 days of incubation at 25°C, single conidial colonies were transferred to potato dextrose agar (PDA; Difco, Detroit, MI, USA). Three isolates were obtained, of which two isolates (accession nos. JBARES74 and JBARES75) were submitted at the herbarium of Jeollabuk-do Agricultural Research and Extension Services, Korea and another isolate (accession no. KACC47790) was deposited at the Korean Agricultural Culture Collection (KACC) of the National Institute of Agricultural Sciences, Korea.
DNA extraction and polymerase chain reaction amplification. Genomic DNA was extracted from two weeks old cultures (KACC47790, JBARES74, and JBARES75) according to the instructions outlined by
Nakashima et al. (2016). Nucleotide sequences of partial actin (
actA), partial translation elongation factor 1-alfa (
tef1), partial DNA-directed RNA polymerase II second largest subunit (
rpb2) genes and internal transcribed spacer (ITS) regions of ribosomal DNA were am-plified using primer sets ACT-512F/ACT-783R, EF1-728F/EF1-986R, fRPB2-5F/fRPB2-7cR, and V9G/ITS4, respectively (Carbone and
Kohn, 1999;
de Hoog and Gerrits van den Ende, 1998;
Liu et al., 1999;
White et al., 1990). The polymerase chain reaction products were sequenced by commercial company Macrogen Inc. (Seoul, Korea) with usage of the same primers. Newly obtained sequences of
actA (221 bp),
tef1 (313 bp),
rpb2 (868 bp) genes and ITS (702 bp) regions were edited in SeqMan software (Lasergene, DNASTAR, Madison, WI, USA) and deposited in GenBank under following accession numbers MW465751, MW465749, MW463367, and MW465750, respectively.
Phylogenetic analyses. Newly obtained sequences were assembled and aligned with 14 sequences belonging to the genus
Pseudocercospora retrieved from the GenBank.
Cercospora cf.
nicotianae (CPC15918) was used as an outgroup. A neighbor-joining (NJ) phylogenetic tree was constructed using the maximum composite likelihood method implemented in MEGA7 (
Kumar et al., 2016). The robustness of the NJ tree was evaluated with 1,000 bootstraps (BS) values.
Pathogenecity test. To conduct a pathogenicity test, totally ten leaves of 1-year-old healthy Chinese fringetree plants (3 per plant) were inoculated with colony discs (∮0.5 cm) obtained from 2-week-old cultures on PDA. As a control treatment, another ten leaves (3 per plant) were inoculated with noncultured discs. To maintain the relative humidity of 100% for 24 hr, treated leaves were covered with plastic bags and kept at open field.
Results
Field obserbations. Leaf spot disease caused by
P. chionanthi-retusi started in August and the disease severity reaches peaks with 100% incidence till November. The fungus attacks older leaves, symptoms appear on both side of the leaves in the form of irregular spots, which then coallasced into big, dark lesions. According to our observations, as the disease progressed, the leaves dried out and prematurely fell off (
Fig. 1A, B).
C. retusus is planted in parks and alongside of roads, fungus reduces the ornamental value of the plant by causing a mess of infected leaf litter.
Fig. 1.
Leaf spot caused by Pseudocercospora chionanthi-retusi on Chionanthus retusus. (A) Leaves were infected, decreasing the vigor of the plants. (B) Infected leaves early dropped due to disease. (C, D) Close-up of symptoms on leaves. (E) Fructification of the fungus on the lesion, and blackish patches showed on the lesion cue to heavy fructification. (F, G) Conidiophores. (H) Conidia. Note: Photomicrographs were taken from voucher specimen KUS-F27686.

Morphology. Leaf spots were angular to irregular, vein-limited, scattered, 1-8 mm diameter, brownish-gray to dark brown when dry, with heavy fructification (
Fig. 1C-E). Sometimes these spots cover half of the leaves when coalesced. Caespituli were amphigenous, but mostly epiphyllous (
Fig. 1E). Mycelium internal, subhyaline to pale brown. Stromata lacking to moderate, composed of a few brown cells while moderately developed, erumpent, epidermal, reaching up to 50 μm diameter on the adaxial leaf surface (
Fig. 1F). Conidiophores were loose to densely fasciculate arising from stromata, occasionally solitary on the creeping hyphae, straight or geniculate, smooth to rough, thin-walled throughout, 0-2 septate, 10-45×2-5 µm (
Fig. 1F, G). Conidia were solitary, holoblastic, hyaline to pale olivaceous, cylindrical to obclavate, smooth, straight to slightly curved, obconically truncate at the base, rounded at the apex, 2-8 septate, 15-52×2-4 µm, with unthickened and not darkened hilum (
Fig. 1H). These morphological characteristics are in well agreement with those reported for
P. chionanthi-retusi (
Nakashima et al., 2016).
Molecular-phylogenetic results. Results of BLASTn search showed that both sequences of partial
tef1 and
actA genes obtained in this study were 100% identical to the sequences KX462671, GU384391 (
tef1), and KX462552 (
actA) of
P. chionanthi-retusi on
C. retusus in GenBank. Sequence for ITS regions showed the highest similarities (>99%) to the sequences of several
Pseudocercospora spp., whereas sequences of rpb2 gene shared 99.85% similarity to the
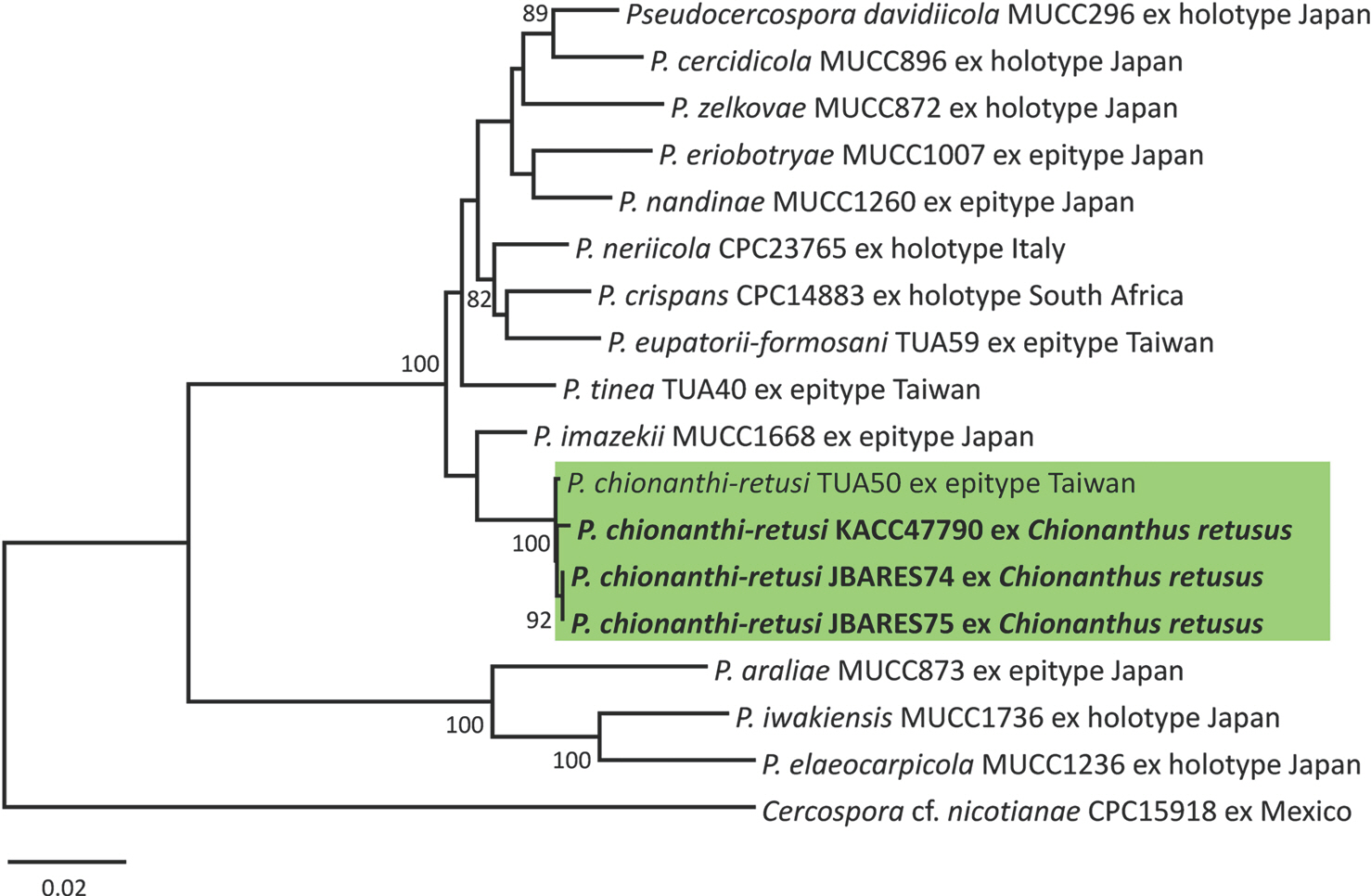
P. chionanthi-retusi (KX462620). As a result of the phylogenetic analysis it was clearly demonstrated that three newly obtained sequences, together with isolates of
P. chionanthi-retusi, formed a separate clade distinct from other
Pseudocercospora species in the phylogenetic tree with a BS support value of 100% (
Fig. 2).
Fig. 2.
Phylogenetic relationship between Pseudocercospora chionanthi-retusi and some reference isolates of Pseudocercospora species retrieved from GenBank, inferred by the neighbor-joining analysis using sequences of the multigene dataset of actin (actA), translation elongation factor 1-alpha (tef1), RNA polymerase II second largest subunit gene (rpb2), and internal transcribed spacer. Bootstrap values (≥80%) are shown on the branches. The Korean isolates used in this study are shown in boldface.
Pathogenecity test result. Typical symptoms of leaf spots appeared on the inoculated leaves 10 days after inoculation and were identical to the symptoms observed in the field. Control leaves remained healthy. P. chionanthi-retusi was reisolated from the lesions of inoculated leaves with confirmation of Koch's postulates.
Discussion
The encounter of
P. chinanthi-retusi on
C. retusus in Korea was reported by
Crous et al. (2013) based on joint research on
Pseudocercospora species. That report refers to a sample that was collected by HD Shin from Osan, Korea (KUS-F17995; 30 Oct 2007) and sent to PW Crous (The Westerdijk Fungal Biodiversity Institute, Netherlands). Obtained isolate was registered as CPC14683 (CPC: Culture collection of Pedro Crous, housed at the Westerdijk Institute). However, a detailed morphological description of
P. chionanthi-retusi was not given in the mentioned study. On the other hand,
Nakashima et al. (2016) provided detailed morphological traits of this fungus, but distribution was confined to Taiwan, Japan, and China. Furthermore, these studies were focused on mycological aspects of fungus, such as taxonomic position and morphology. Thus, in this study, we aimed to highlight phytopathogenic value of
P. chinanthi-retusi for future studies with the provision of morphological and molecular-phylogenetic data of this species on
C. retusus based on Korean isolates. As mentioned before this pathogen causes early defoliation of plant and having regard cold winter in Korea, it is highly likely that the
C. retusus will suffer frost damage during the winter season. Taking into account the economic value of this fungus, it is necessary to develop an effective control technology against this pathogen.
Acknowledgments
This study was carried out with the support of the “Cooperative Research Program for Agricultural Science & Technology Development (Project Nos. PJ014976)” Rural Development Administration, Republic of Korea.
REFERENCES
Carbone, I. and Kohn, L. M. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes.
Mycologia 91: 553-556.

Crous, P. W., Braun, U., Hunter, G. C., Wingfield, M. J., Verkley, G. J., Shin, H. D. et al. 2013. Phylogenetic lineages in
Pseudocercospora.
Stud. Mycol. 75: 37-114.



de Hoog, G. S. and Gerrits van den Ende, A. H. G. 1998. Molecular diagnostics of clinical strains of filamentous Basidiomycetes.
Mycoses 41: 183-189.


Liu, Y. J., Whelen, S. and Hall, B. D. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit.
Mol. Biol. Evol. 16: 1799-1808.


Nakashima, C., Motohashi, K., Chen, C.-Y., Groenewald, J. Z. and Crous, P. W. 2016. Species diversity of
Pseudocercospora from Far East Asia.
Mycol. Prog. 15: 1093-1117.


White, T. J., Bruns, T., Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a Guide to Methods and Applications, eds. by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, pp. 315-322. Academic Press, New York, NY, USA.












