Colletotrichum aenigma Associated with Apple Bitter Rot on Newly Bred cv. RubyS Apple
Article information
Abstract
The abnormal brown sunken lesions were observed on cv. RubyS apple fruits in an orchard located in Gunwi, Gyeongbuk province, Korea. The primary observed symptoms such as small round sunken lesions and small black dots on the symptomatic area were different from the reported apple diseases. The affected apple fruits were sampled and subjected to isolation of the causal agent. Cultural and morphological characteristics of isolated fungal strain, designated KNUF-20GWA4, were similar to that of Colletotrichum spp. Based on multilocus sequence analyses using internal transcribed spacer regions and partial sequences of β-tubulin, glyceraldehyde-3-phosphate dehydrogenase, chitin synthase, and actin genes, strain KNUF-20GWA4 showed 99.2-100% similarities with C. aenigma ICMP 18608 and the isolate clustered together with several other strains of this species in the phylogenetic tree. To our knowledge, this is the first report of bitter rot on apple fruits caused by C. aenigma.
Apples (Malus domestica) are cultivated worldwide, with production reaching approximately 87 million tons in 2019 (FAOSTAT, 2019). In Korea, the area used for apple cultivation is approximately 30,000 ha, and about 90% of apple cultivars are Fuji and Hongro (Choi et al., 2018). A new apple cultivar, ‘RubyS’, was developed by the National Institute of Horticultural Science at the Rural Development Administration and registered in 2015. The main characteristics of cv. RubyS are high resistance to apple bitter rot and longer storability at room temperature than other apple cultivars (Kwon et al., 2019). Due to the demand for medium-sized apples in Korea, the cv. RubyS cultivation area has increased to approximately 105 ha. The damage caused by various fungal diseases is considered to be one of the main factors limiting the commercial production of apples. It is well known that the key to effective disease management starts with the correct diagnosis of the causal agent. In this study, a fungal pathogen was isolated from the affected cv. RubyS apple fruits and it was identified as a new causal agent of apple bitter rot in Korea.
In September 2020, abnormal brown spot symptoms were observed on cv. RubyS apple fruits in an orchard located in Gunwi, Gyeongbuk province, Korea. The estimated infection rate was observed around 5% on the surveyed orchard. The affected areas were light to dark brown inside, surrounded a dark ring. A solid symptomatic lesion, circular or irregular in shape, and small black dots were observed on the symptomatic area (Fig. 1A–D). These symptomatic features were distinct from the symptoms observed on infected apple fruits that are typical of fungal diseases such as bitter rot or white rot (Uhm, 2010). The causal agent was isolated from the abnormal brown spots by wiping the surface of an affected apple with 70% EtOH. Then, a thin layer of the diseased peel was removed using a sterilized blade, and the diseased tissues were directly transferred onto potato dextrose agar (PDA; Difco, Detroit, MI, USA) and incubated at 25oC. After 3 days, white mycelium was transferred onto new PDA plates and formed colonies that were used to further identify the strain, designated KNUF-20GWA4. The fungal isolate was identified by extracting genomic DNA from a 5-day-old fungal culture grown on PDA medium using a HiGeneGenomic DNA Prep Kit (BIOFACT, Daejeon, Korea), as per the manufacturer’s protocol. The internal transcribed spacer (ITS) regions, including ITS1, 5.8S, and ITS2, were amplified using the ITS1/ ITS4 (White et al., 1990). The amplified PCR products were purified with ExoSAP-IT PCR Product Cleaning Reagent (Thermo Fisher Scientific, Waltham, MA, USA) then requested the sequencing analysis (SolGent, Daejeon, Korea). A sequence of 581 bp was obtained from the ITS regions. A BLAST search in the National Center for Biotechnology Information database revealed similarities of 100% between the ITS regions of KNUF-20GWA4 and several strains belonging to Colletotrichum aenigma, C. siamense, and C. gloeosporioides, indicating that the ITS sequence was unsuitable for precise identification of the isolate.

Abnormal brown spot symptoms observed on cv. RubyS apple fruits and morphological characteristics of the isolate. (A–D) Photographs showing abnormal brown sunken lesion on RubyS diseased apple fruits collected from Gunwi in 2020. (A) Infected RubyS apple collected from the field. (B) Close-up view of the infected apple in A. (C) Symptomatic area on a RubyS apple. (D) Black dots on the symptomatic region. (E) Five-day-old fungal culture on potato dextrose agar (PDA). (F–I) Light microscopy showing conidia (F), conidiomata (G), a conidiogenous cell (H), and appressoria (I) from the PDA culture. Scale bars=10 μm (F–I).
Recently, various combinations of molecular markers were used for multilocus phylogenetic analysis to distinguish closely related taxa in C. gloeosporioides species complexes (Fu et al., 2019; Oo et al., 2017; Wang et al., 2021). Thus, we used this approach to amplify the β-tubulin (β-TUB), glyceraldehyde-3-phosphate dehydrogenase (sphate dehydrogenase), chitin synthase (CHS), and actin (ACT) genes of strain KNUF-20GWA4 according to previous reports (Carbone and Kohn, 1999; Glass and Donaldson, 1995; Guerber et al., 2003). The amplification of β-TUB, sphate dehydrogenase, CHS, and ACT loci yielded fragments of 580, 266, 270, and 254 bp, respectively. All the gene sequences were registered in NCBI with the following GenBank (accession numbers MZ068034, LC630937–LC630940, respectively.). Based on the partial gene sequences of the four molecular markers, strain KNUF-20GWA4 showed 99.2–100% similarities with those of C. aenigma ICMP 18608T. To confirm the closest relationship, phylogenetic analysis using concatenated sequences of the ITS regions, β-TUB, sphate dehydrogenase, CHS, and ACT genes were performed. The sequences of allied species were retrieved from NCBI GenBank (Table 1) and a phylogenetic tree was constructed using the maximum-likelihood method as implemented in MEGA7 software (Kumar et al., 2016). Strain KNUF-20GWA4 clustered together with C. aenigma IY1059, C. aenigma GRAP12, and C. aenigma GRAP07 isolated in Korea (Choi et al., 2017; Kim et al., 2021) and two other strains of C. aenigma ICMP 18608 and ICMP 18686 in a monophyletic clade with a high bootstrap value, strongly supporting their affiliation to the same species (Fig. 2).
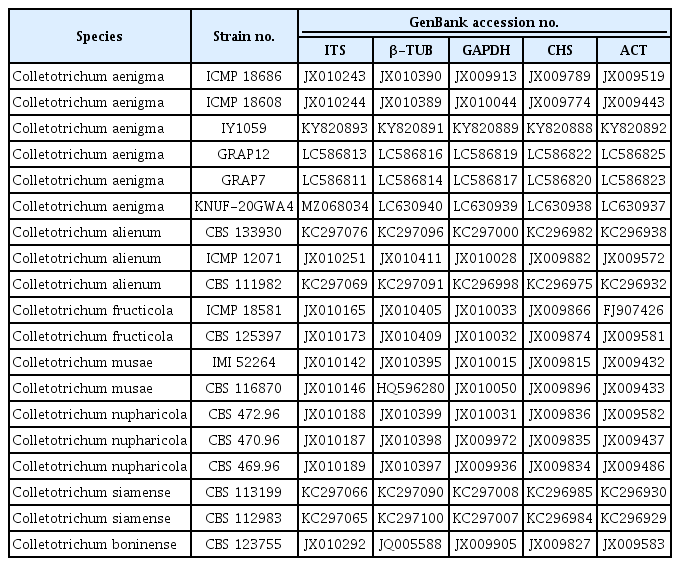
Colletotrichum species used in this study for phylogenetic analysis and their GenBank accession numbers

Maximum-likelihood phylogenetic tree, based on the concatenated sequences (ITS+β-TUB+GAPDH+CHS-1+ACT), shows the phylogenetic position of Colletotrichum aenigma KNUF-20GW4 among the closest Colletotrichum species. The isolated strain is shown in bold. Bootstrap values (based on 1,000 replications) >70% are shown at the branch points. Colletotrichum boninense CBS 123755 was used as the outgroup. Scale bar=0.02 substitutions per nucleotide position.
Strain KNUF-20GWA4 was cultured for 7 days on PDA at 25oC, and colony characteristics, such as color, shape, and size, were recorded. The colonies were floccose and white to gray with a white margin, with a dark green to gray center on reverse. The morphological characteristics were observed by light microscopy using a BX50 microscope (Olympus, Tokyo, Japan). The conidia were abundant and cylindrical with round ends, sometimes slightly curved, tapering toward 1 or both ends, and measured 14.9–21.1×5.7–8.5 μm (mean, 18.6×7.3; n=50) The appressoria were brown to dark brown, clavate, crenate with lobes, and measured 9.2–17.7×5.2–8.8 μm (mean, 12.0×7.5; n=30) (Fig. 1E–I). These morphological and cultural characteristics of isolate KNUF-20GWA4 were in good agreement with those previously reported for C. aenigma (Wang et al., 2016; Weir et al., 2012), thus strongly supporting phylogenetic analysis results of KNUF-20GWA4 based on the above-mentioned five concatenated sequences.
Strain KNUF-20GWA4 was inoculated onto three healthy cv. RubyS apple fruits to confirm its pathogenicity. The spore suspension for the inoculum was prepared after culturing the isolate for 15 days, and the conidial concentration was determined with a hemocytometer and adjusted to approximately 6.8×105 conidia/ml. The surface of a healthy apple fruit was wiped using 70% EtOH, and two lenticels were wounded using a sterilized needle. Then, paper disks containing 30 μl of spore suspension were attached and sealed using foil, while sterilized water was used as a mock inoculation. Identical paper disks were also attached to unwounded lenticels and sealed using foil. After 14 days, brown spots with conidiomata were only observed on the wounded points on all the inoculated fruits, while no symptoms were observed on the unwounded and mock-inoculated parts (Fig. 3). The symptomatic lesions slowly increased in size and were solid, similar to the natural disease symptoms. The fungal strain was re-isolated and identified as C. aenigma from the symptomatic areas of each inoculated fruit (data not shown). Meanwhile, C. aenigma KNUF-20GWA4 was also inoculated on cv. Fuji apple fruits using the same method, however, no symptoms appeared after 14 days (data not shown).

Pathogenicity test results of Colletotrichum aenigma KNUF-20GWA4. (A, B) Symptoms on the inoculated cv. RubyS apple. (C) Enlarged picture of B showing the conidiomata on the symptomatic area.
C. aenigma has been reported as a pathogen associated with anthracnose on several hosts, including grape and Sedum kamtschaticum (Orange stonecrop) in Korea (Choi et al., 2017; Kim et al., 2021); grapevine, Asian pear, and walnut in China (Fu et al., 2019; Wang et al., 2021; Yan et al. 2015); avocado in Israel (Sharma et al., 2017); Synsepalum dulcificum (miracle fruit) in Japan (Truong et al., 2018); and Hylocereus undatus (dragon fruit) in Thailand (Meetum et al., 2015). However, this fungal pathogen has not been reported as the causal agent of bitter rot on apple fruits.
Until now, various Colletotrichum species have been reported on the apple fruits in Korea and causes significant economic losses in apple cultivation (Oo et al., 2018; Uhm, 2010). Although it was confirmed that the isolated C. aenigma has no pathogenicity on cv. Fuji apples in this study, however, it is necessary to confirm the pathogenicity on the different apple cultivars and the possibility of co-infection. To our knowledge, this is the first report of C. aenigma as the agent of apple bitter rot on apple cv. RubyS. Further studies are necessary to better define the host range of C. aenigma in Korea, the impact of the disease on yield, and control methods.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This research was supported by Kyungpook National University Research Fund, 2018.
