First Report of Botryosphaeria parva Causing Stem Blight on Rubus crataegifolius in Korea
Article information
Abstract
In 2015, stem blight of Rubus crataegifolius was observed in Pohang, Korea. The symptoms began as dark red spots in the stem, which led to stem blight, then leaf blight, and eventually resulted in death. A fungal isolate was obtained from a symptomatic stem and incubated on a potato dextrose agar plate. The isolated fungus produced white, cloudy mycelia turned black in 3 days. Based on the morphological characteristics, the causal fungus was assumed to be Botryosphaeria sp. A pathogenicity test was conducted according to Koch's postulates. To identify the causal agent, the combined sequence of the internal transcribed spacer, β-tubulin, and translation elongation factor 1α genes were used for phylogenetic analysis. Approximately 1,200 bp of the combined sequence clearly suggested that the isolated pathogen was Botryosphaeria parva. This is the first report on stem blight in R. crataegifolius caused by B. parva in Korea.
Body
Rubus crataegifolius Bunge belongs to the family Rosaceae, which is commonly found in the mountainous areas of Korea, Japan, and northeast China. R. crataegifolius has been used as food and traditional medicine for many years in this region by its various bioactivities such as immunological and antioxidant activities (Moon et al., 2011; Ni et al., 2009; Zhang et al., 2010). According to the usefulness of R. crataegifolius, the importance of disease management of R. crataegifolius has increased in Korea (Korea Forest Service, 2015). Several pathogens that infect R. crataegifolius have been reported in Korea, including Septoria rubi, Rhizopus stolonifer, Phragmidium griseum, and Phytoplasma (Kim et al., 2009). In July 2015, stem blight symptoms were observed in a field of R. crataegifolius in Pohang, Gyeongbuk province, Korea. Early in the course of the disease, plants showed dark red spots on stems; these spots expanded to black lesions associated with stem blight (Fig. 1A). As the disease progressed, pycnidia appeared on the stem lesions (Fig. 1B). The blight eventually spread to the leaves and resulted in plant death.
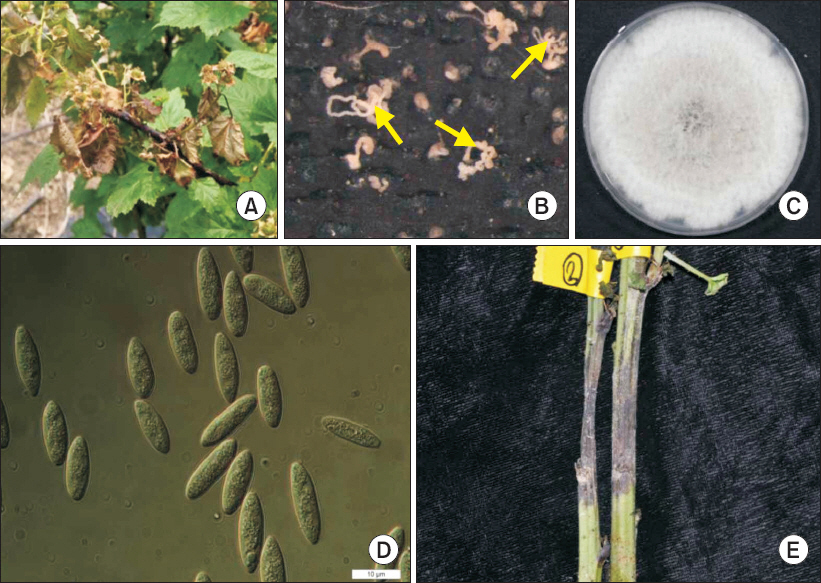
(A) Symptoms of stem blight on Rubus crataegifolius. (B) Conidia discharged from pycnidia on the lesion (yellow arrows). (C) Seven-day-old fungal isolate on potato dextrose agar medium. (D) Conidia. Scale bar=10 μm. (E) Symptoms following experimental inoculation.
A fungus, the presumed causal agent, was isolated from the diseased plant and designated as PPL02. The infected part of the stem was cut into small pieces and the surface of pieces was sterilized with 1% NaOCl for 1 minute and washed three times with 70% ethanol. The surface-sterilized pieces were placed onto potato dextrose agar (PDA) medium and incubated at 25°C. For isolation of a pure culture, hyphal tips of mycelia on the PDA medium were cut and transferred to fresh PDA plates. The isolated pathogen PPL02 produced white, cloudy mycelia that reached the edge of the PDA plate within 7 days (Fig. 1C). The center of the colony turned black after 3 days of incubation, with several dark spots appearing on the back side of the plate, and the black part extended all over the plate within 21 days of incubation. When the fungal colony was exposed to visible light with a 12-hour photoperiod for 21 days, the fungus produced conidia, which were aseptate, hyaline, and ellipsoid, ranging from 13–18 μm×4–6 μm (Fig. 1D). The morphology of the colonies and conidia showed similar characteristics to Botryosphaeria species. Although the culture characteristics and morphologies of B. parva and B. dothidea are similar, guidelines to distinguish these species have been previously suggested (Phillips, 2002). According to the previous report, conidia of B. parva have 14 to 18 μm length with length to width (L/W) ratio 3.2 to 3.9 while conidia of B. dothidea have 20 to 28 μm length with L/W ratio 4.3 to 5.2 (Phillips, 2002). Previously reported B. parva isolates from various hosts have satisfied these criteria (Haleem et al., 2012; Linaldeddu et al., 2007; Phillips, 2002; Slippers et al., 2005). Thus, morphological and cultural observation suggested that PPL02 was presumed to be B. parva, based on its morphological and culture characteristics (Table 1).

Morphological comparision of the isolate obtained from Rubus crataegifolius Bunge and of Botryosphaeria species described previously
To verify pathogenicity of PPL02, a fungal disc was obtained from the PDA plate at 7 days of incubation. The fungal disc was attached to an artificially wounded stem of a healthy plant of mature R. crataegifolius and then was wrapped with parafilm to maintain humidity. Ten days after inoculation, the same symptoms that were observed in the original plant appeared on the inoculated stem (Fig. 1E). Re-isolated pathogen from the disease lesion on the inoculated stem exhibited the same morphological characteristics of the original isolate. Therefore, PPL02 fulfilled Koch's postulates for establishing the causative agent of blight on R. crataegifolius.
A phylogenetic analysis was performed to clarify the species of this causal agent of stem blight in R. crataegifolius. Genomic DNA was extracted from PPL02, grown on the PDA plate, using a HigeneTM Genomic DNA Prep Kit (Biofact, Daejeon, Korea). PCR was performed with genomic DNA to amplify the internal transcribed spacer (ITS) region, β-tubulin, and translation elongation factor 1α (EF1-α) genes. Since some species belonging to Botryosphaeria genus are indistinguishable in the phylogenetic analysis using ITS only (Slippers et al., 2004), in this study, we used three genes and its combined sequences for further analysis. Each gene was amplified with the following, respective primer sets: ITS1F/ITS4 for ITS region (White et al., 1990), Bt2a/Bt2B for β-tubulin (Glass and Donaldson, 1995), and EF1-728F/EF1-986R for EF1-α (Carbone and Kohn, 1999). The sequence of each gene was compared with sequences in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST) by BLAST search. Nucleotide sequences of each gene showed >99% similarity with sequences of B. parva (accession Nos. EU938334, FJ238525, DQ487158). Analyzed sequence data were deposited in GenBank (accession Nos. LC120826 for ITS region, LC120827 for β-tubulin, and LC120828 for EF1-α). For the phylogenetic analysis, reference genes of various species belonging to the genus Botryosphaeria were obtained from GenBank (Table 2). Sequences of three genes for each species, including the fungal isolate characterized in this study, combined as follows: ITS region, β-tubulin, and EF1-α. Combined sequences were aligned and analyzed using analysis tool provided in the DDBJ (DNA databank of Japan) homepage (http://www.ddbj.nig.ac.jp/). A phylogenetic tree was constructed using TreeView program with the combined sequences (Page, 1996). A results showed that our combined sequence was clustered with B. parva sequences (Fig. 2). Taken togather, the pathogen causing stem blight on R. crataegifolius was identified as B. parva, based on morphological and phylogenetic analyses.
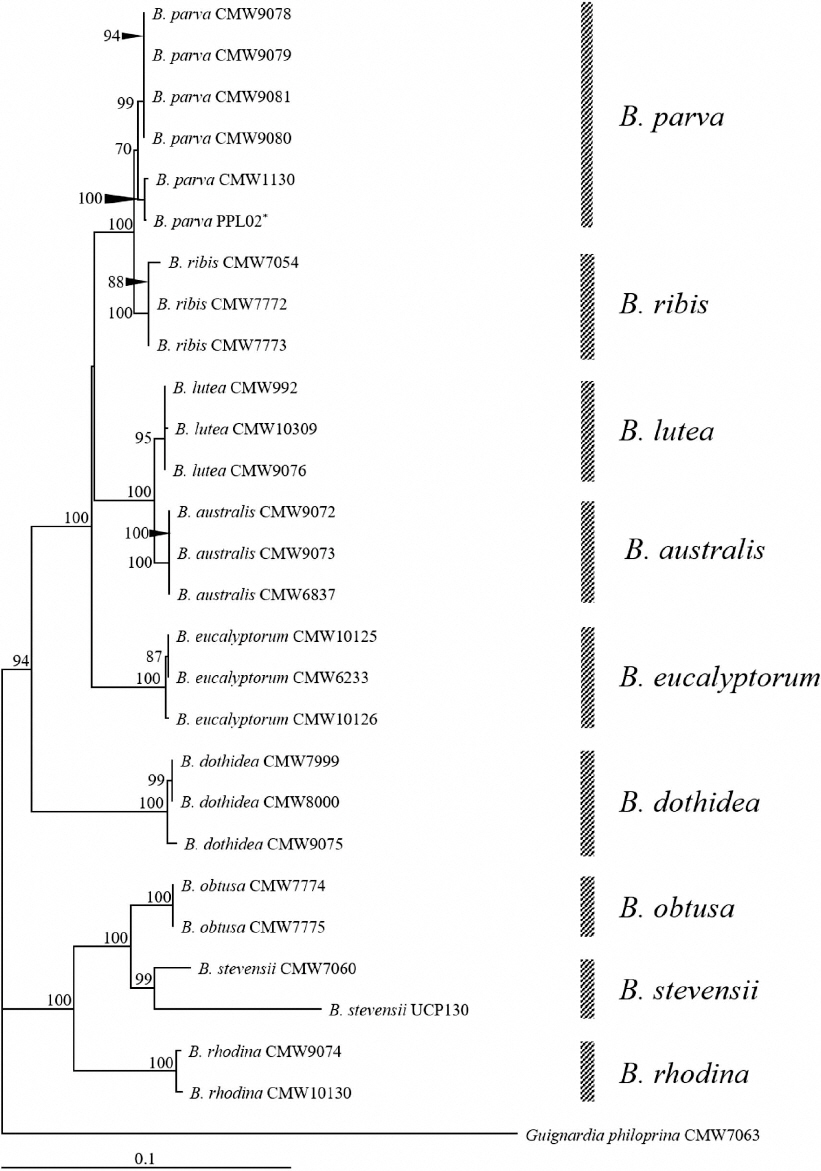
Phylogenetic analysis of Botryosphaeria species using a combined sequence of the internal transcribed spacer, β-tubulin, and elongation factor 1α genes. The tree was constructed using the neighbor-joining method with 1,000 replicates.
*The fungus isolated in this study.
B. parva was first reported to cause soft rot on kiwifruit in New Zealand (Pennycook and Samuels, 1985). Since this first report, presence of B. parva has been reported in 71 host species across six continents and 21 countries (Sakalidis et al., 2013). This pathogen causes serious symptoms, including dieback, canker, stem blight, and fruit rot in various hosts, such as grapevines, kiwi vines, cherry trees, raspberry bushes, and mango (Abdollahzadeh et al., 2013; Haleem et al., 2012; Linaldeddu et al., 2007; Pennycook and Samuels, 1985; Phillips, 2002; Sakalidis et al., 2013; Slippers et al., 2005; van Niekerk et al., 2004). Thus, the characterization and management of this pathogen is very important. In Korea, B. parva was not reported until 2009 (Kim et al., 2009). In 2012 and 2013, dieback symptoms caused by Neofusicoccum parvum, an anamorphic synonym of B. parva, on blueberries and walnuts has been reported (Cheon et al., 2013; Choi et al., 2012), but there has been no report hitherto that B. parva (N. parvum) was isolated from R. crataegifolius. Thus, this is the first report of stem blight on R. crataegifolius caused by B. parva in Korea. Considering economic value of R. crataegifolius, methods to manage this pathogen are urgently needed to prevent low production of R. crataegifolius.
Acknowledgements
This research was supported by a grant (NIBR 2015-01205) from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea for projects on the survey and discovery of indigenous Korean fungal species.
