Genome-Based Insights into the Thermotolerant Adaptations of Neobacillus endophyticus BRMEA1T
Article information
Abstract
The bacterium Neobacillus endophyticus BRMEA1T, isolated from the medicinal plant Selaginella involvens, known as its thermotolerant can grow at 50°C. To explore the genetic basis for its heat tolerance response and its potential for producing valuable natural compounds, the genomes of two thermotolerant and four mesophilic strains in the genus Neobacillus were analyzed using a bioinformatic software platform. The whole genome was annotated using RAST SEED and OrthVenn2, with a focus on identifying potential heat-tolerance-related genes. N. endophyticus BRMEA1T was found to possess more stress response genes compared to other mesophilic members of the genus, and it was the only strain that had genes for the synthesis of osmoregulated periplasmic glucans. This study sheds light on the potential value of N. endophyticus BRMEA1T, as it reveals the mechanism of heat resistance and the application of secondary metabolites produced by this bacterium through whole-genome sequencing and comparative analysis.
Selaginella involvens is a medicinal plant that has been used for the prevention and treatment of asthma, yet it has received little attention in scientific research (Qiu et al., 2020). However, this plant is rich in secondary metabolites, including phenol-based compounds (such as flavonoids, tannins, and saponins), terpenoids (such as triterpenes), steroids, alkaloids, selaginellin, and sumaflavone (Abeysinghe et al., 2021; Adnan et al., 2021; Li et al., 2021; Woo et al., 2021). These compounds possess a range of important biological activities, including antioxidant, anti-allergic, anticancer, antimicrobial, anti-inflammatory, antifungal, antibacterial, UV irradiation-protective, antihypertensive, anticlotting, and an-ti-acne properties (Joo et al., 2008; Lei et al., 2021; Setyawan and Darusman, 2008). The Selaginella species, including Selaginella involvens, are well known for their unique ability to tolerate desiccation, which refers to the ability of plants, animals, and microbiomes to tolerate and survive extremely dry conditions. Endophytes, which are microorganisms that inhabit the interior tissues of plants, provide beneficial functions to their host plants, for example, in enhancing a host plant's resistance to both biotic and abiotic stresses by secreting secondary metabolites, such as phytohormones that promote plant growth, or enzymes that activate the plant immune system to protect against phytopathogens (Baek et al., 2019; Etesami and Beattie, 2018; Hingston et al., 2017; Kumar et al., 2020; Sunita et al., 2020). Endophytes are known to be rich sources of various novel cytotoxic compounds, including antibacterial substances, insecticides, an-ticarcinogenic molecules, and biostimulants for essential oil biosynthesis. These compounds hold significant commercial value and find wide applications in the medical, agricultural, and cosmetic industries (Gupta et al., 2020; Li et al., 2018; Wang et al., 2006; Zhao et al., 2011). Endophytes can also play a crucial role in promoting the production of secondary metabolites in their host plants. They do so by cooperating with the plant to complete signal pathways, similar to key enzyme biosynthesis pathways, or by influencing the direction of certain reactions in the host's metabolism to produce specific metabolites (Qadri et al., 2017; Zhao et al., 2011). Since medicinal plants are a valuable source of therapeutic compounds, and since the secondary metabolites are synthesized by symbiotic interactions, it is important to isolate such compounds from the endophytes associated with these plants. In our investigation of the endophytic bacterial diversity of Selaginella involvens, we identified a novel species of the genus Neobacillus, which we named Neobacillus endophyticus BRMEA1T. This Gram-positive, endospore-forming bacterium was found to be thermotolerant and capable of growing at a temperature up to 50°C (Jiang et al., 2019). The genus Neobacillus was reclassified from Bacillus in 2020, and as of March 2023, there are 22 validly published species in this genus. Some members of Neobacillus are thermotolerant, and can grow at temperatures up to 60°C (Han et al., 2013; Heyrman et al., 2004). To improve our understanding of how these endophytes adapt to high temperatures, we sequenced the entire genome of Neobacillus endophyticus BRMEA1T and analyzed its potential heat-tolerance-related genes by comparing the entire genomes of thermotolerant strains with mesophilic strains within the same genus.
Genomic DNA was extracted and sequenced as previously described (Jiang et al., 2019). We utilized both Illumina (San Diego, CA, USA) and PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) platforms to perform whole-genome sequencing (WGS) of N. endophyticus BRMEAT. The complete genome of N. endophyticus BRMEAT comprised three contigs with a total of 5,632,809 base pairs, with contig lengths of 5,239,118, 356,662, and 37,029 base pairs, respectively. Annotation of the genome was performed by NCBI PGAP using GeneMarkS-2+ (V.4.11), based on the best-placed reference protein set. The genome contained 5,291 protein-coding genes, 186 transfer RNA (tRNA) genes, 51 ribosomal RNA (rRNA) genes, 6 non-coding RNA (ncRNA), and 107 pseudogenes (Supplementary Table 1). No plasmid sequences were detected from the whole genome. Analyses with VirulenceFinder and ResFinder revealed the absence of known virulence and antibiotic resistance genes, suggesting that N. endophyticus BRMEA1T could be generally recognized as a safe (GRAS) strain, similar to other members of the Bacillus genus that can be safely used in various industries and agricultural applications.
The entire genome of BRMEA1T was uploaded to the TYGS website, and all of the type strains in the genus Neobacillus were selected for taxonomy-based identification. Strain BRMEA1T and all of the type strains were pairwise compared for accurate intergenomic distances under the “trimming” algorithm and distance formula d5. At the same time, the digital DNA-DNA hybridization (dDDH) values between BRMEA1T and all of the type strains were calculated with the recommended settings of GGDC (Meier-Kolthoff and Göker, 2019). The minimum evolution tree was created via FASTME 2.1.4, with 100 pseudo-bootstrap replicates for branch support. The genome comparisons between the members in genus Neobacillus were performed, and a heatmap based on the OrthoANI values was calculated with the Orthologous Average Nucleotide Identity Tool (OAT) (Lee et al., 2016). A comparison and annotation with clusters of orthologous groups (COGs) were performed using OrthoVenn2 among thermotolerant strains N. endophyticus BRMEA1T (50°C) and N. drentensis DSM 15600T (=NBRC 102427T; 50°C), and four mesophilic strains including N. cucumis DSM 101566T (45°C), N. mesonae FJAT-13985T (45°C), N. niacini DSM 2923T (=NBRC15566T; 40°C), and N. notoginsengisoli SYP-B691T (42°C). To visualize the coding sequence identities among the closest related strains in the genus Neobacillus, we utilized the BLAST ring image generator (BRIG) software with the BLASTn algorithm (Alikhan et al., 2011).
To determine the taxonomic position of N. endophyticus BRMEA1T within the genus Neobacillus, a whole-genome-based phylogenetic tree was constructed using the type strains of all of the species in the genus via the TYGS server (Supplementary Fig. 1). N. endophyticus BRMEA1T formed a cluster with N. fumarioli NBRC 102428T, which was consistent with a previous study (Jiang et al., 2019). However, the ANI and dDDH values were only performed between strain BRMEA1T and four type strains in the genus Neobacillus, which showed that the ANI value was below 74.8% and the dDDH values were lower than 22.0%. In the present study, we performed the ANI, OrthoANI, and dDDH values based on the whole genome (Supplementary Fig. 2, 3) for all type strains in the genus Neobacillus, results showed that all of the values were below the proposed cutoff values for species delineation (<70% for dDDH and <95% for ANI) (Chun et al., 2018). The genome size of the Neobacillus species ranged from 3,208,840 bp to 6,183,708 bp, with G+C content ranging from 37.4% to 43.8%, and the number of protein-coding genes ranging from 3,025 to 5,944 (https://www.ncbi.nlm.nih. gov/genome/?term=neobacillus). The genome properties of N. endophyticus BRMEA1T fell within the range of these values, indicating that the genome of N. endophyticus BRMEA1T is highly conserved (Supplementary Table 1).
The complete genome sequence of N. endophyticus BRMEA1T underwent annotation using RAST (http://rast.nmpdr.org/). The subsystem coverage was 21%, with a total of 1,259 genes in the subsystem. A total of 4,786 genes were not part of a subsystem, including 1,651 non-hypothetical and 3,135 hypothetical coding DNA sequences (CDSs). Annotation results from the RAST viewer showed that 52 genes were involved in stress response, including 3 genes in osmotic stress (2 in osmoregulation, 1 in the synthesis of osmoregulated periplasmic glucans), 21 in oxidative stress (2 in protection from reactive oxygen species, 14 in oxidative stress, 4 in glutathione, 1 in glutaredoxins), 7 in detoxification (1 in the uptake of selenate and selenite), 26 in general stress response (2 in sigmaB stress response regulation, 3 in dimethylarginine metabolism, 18 in bacterial hemoglobins, 2 in hfl operon, and 1 in carbon starvation), and 1 in periplasmic stress response (Table 1, Supplementary Fig. 4).
Next, we investigated the genes responsible for the thermotolerance ability of N. endophyticus BRMEA1T. We generated a genome comparison map using BRIG to compare two thermotolerant members N. endophyticus BRMEA1T (50°C) and N. drentensis DSM 15600T (50°C), with four mesophilic members of the genus Neobacillus (N. cucumis DSM 101566T [45°C], N. mesonae FJAT-13985T [45°C], N. niacini DSM 2923T [40°C], and N. notoginsengisoli SYP-B691T [42°C]) (Fig. 1). The entire genome of the six type strains was annotated using the RAST SEED server, one user-friendly website that provides access of high-throughput, high-quality annotation for the prokaryotic genome. The results showed that the thermotolerance bacteria N. endophyticus BRMEA1T and N. drentensis DSM 15600T have 52 and 48 genes related to multi-stress response, respectively, and the other four mesophilic members in genus Neobacillus have 46, 45, 45, and 24 genes involved in stress response (Table 1). The osmoregulation, oxidative stress, bacterial hemoglobins, and hfl operon genes have relatively more numbers than the mesophilic strains, N. cucumis DSM 101566T, N. mesonae FJAT-13985T, N. niacini DSM 2923T, and N. notoginsengisoli SYP-B691T (Table 1, Supplementary Table 2).
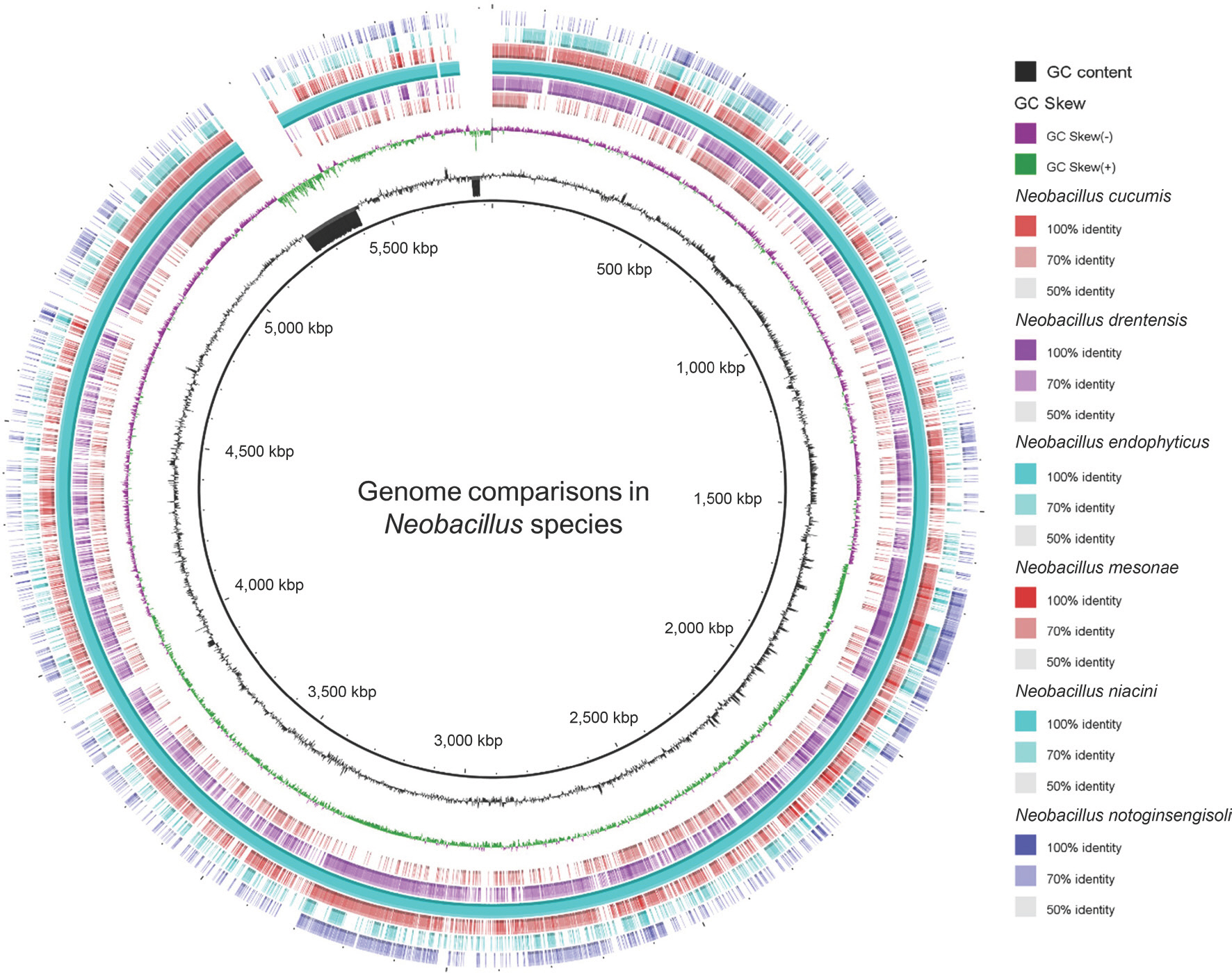
Comparative genome analysis of Neobacillus species. The circular genomic map was created using BRIG v0.95, a software tool for comparative genomics. The map includes several features depicted in different circles: circle 1 represents the genome size, circle 2 represents the GC content, circle 3 represents the GC skew, and circles 4 to 8 represent the comparative genomic maps of two thermotolerant strains (N. endophyticus BRMEA1T and N. drentensis DSM 15600T) and four mesophilic strains (N. cucumis DSM 101566T, N. mesonae FJAT-13985T, N. niacini NBRC 15566T, and N. notoginsengisoli JCM 30743T).
The initial assembled sequences of N. endophyticus BRMEA1T were evaluated using the COGs, a collection of genes that are inherited from a common ancestor across different species. Among the assigned COGs, 28.8% were categorized as unknown functions, while COGs involved in amino acid transport and metabolism (6.8%) and transcription (6.7%) were highly represented in the entire genome (Supplementary Table 3, Supplementary Fig. 5). Notably, amino acids that regulate nitrogen transport and response to abiotic stress (Wan et al., 2017), nitrogen regulatory protein genes (yaaQ, glnB, ntcA_1, and ntcA_2), and hydrogenase/urease maturation factor Hyp were present in the genome (Baillo et al., 2019). These findings suggest that N. endophyticus BRMEA1T may play a crucial role in nitrogen transport and the response to abiotic stress. Furthermore, abiotic stress in plants is known to adversely affect plant growth and yield due to environmental conditions such as salt, drought, cold, high temperature, heavy metals, and UV radiation. Interestingly, genes related to amino acid transport and metabolism accounted for a significant proportion (6.8%) of the total genome of BRMEA1T, suggesting that they may be involved in the response to abiotic stress.
To improve understanding of the relationships of gene function and taxonomic classification between N. endophyticus BRMEA1T and closely related species of the genus Neobacillus via whole genomes, the online program OrthoVenn2 was used to compare genomes between two thermotolerant strains, N. endophyticus BRMEA1T (50°C) and N. drentensis DSM 15600T (50°C), and four mesophilic strains, N. cucumis DSM 101566T (45°C), N. mesonae FJAT-13985T (45°C), N. niacini DSM 2923T (40°C), and N. notoginsengisoli SYP-B691T (42°C). N. endophyticus BRMEA1T and N. drentensis DSM 15600T had 5,178 and 4,920 proteins, and 3,633 and 3,964 gene clusters, respectively, while the mesophilic group members had 5,381, 5,350, 1,922, and 4,594 proteins, and 4,125, 4,144, 1,594, and 3,311 gene clusters, respectively. Eighty-seven genes were unique to both thermotolerant strains (Fig. 2), and were annotated to have functions in biological processes (16), metabolic processes (10), cellular metabolic processes (9), cellular processes (12), nitrogen compound metabolic processes (8), and others (Supplementary Table 3). N. endophyticus BRMEA1T had 128 unique protein clusters, while N. drentensis DSM 15600T had 28 unique protein clusters (Fig. 2, Supplementary Table 4).
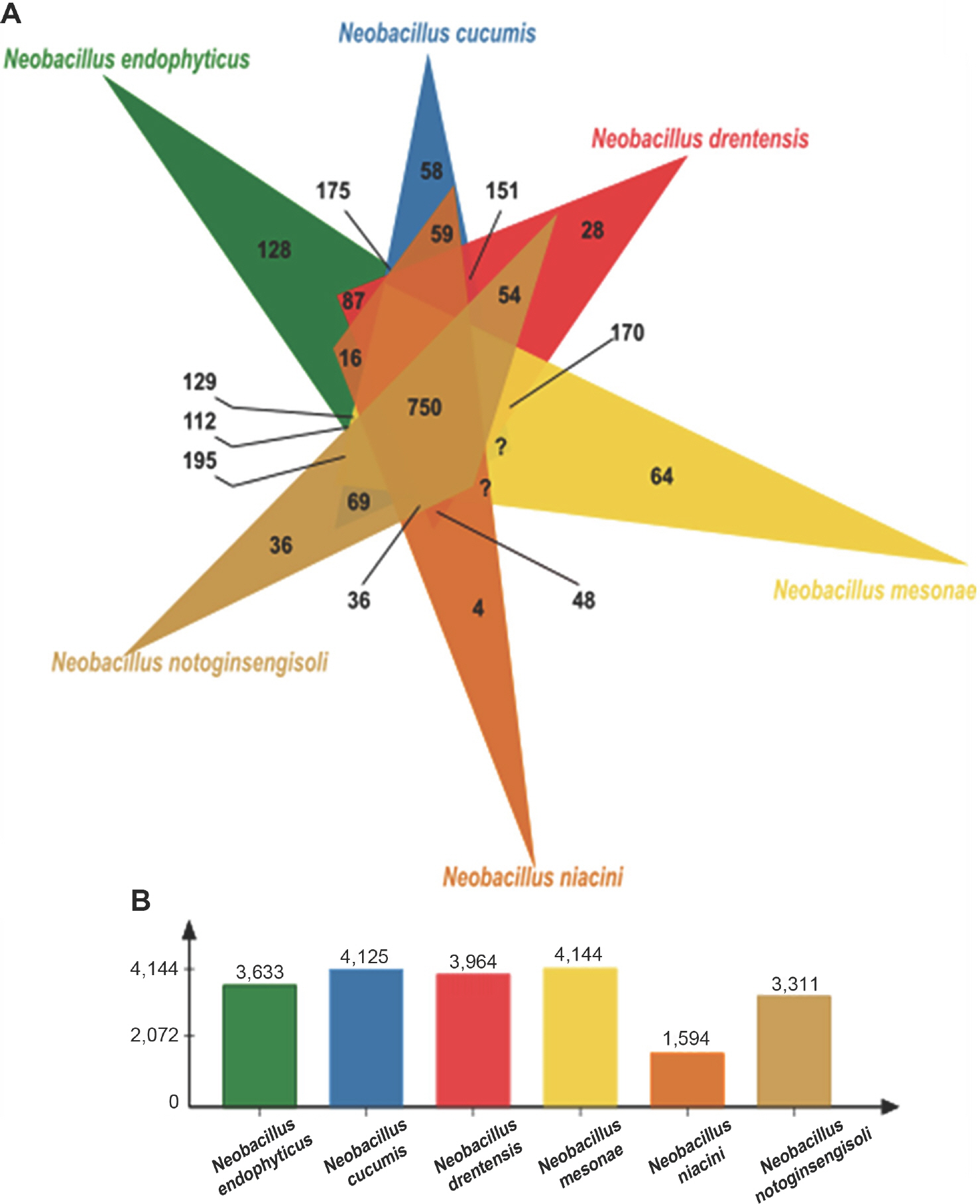
Comparative genomics showing shared and unique orthologous gene clusters in Neobacillus strains. The strains analyzed in this study include two thermotolerant strains, N. endophyticus BRMEA1T and N. drentensis DSM 15600T, as well as four mesophilic strains, N. cucumis DSM 101566T, N. mesonae FJAT-13985T, N. niacini NBRC 15566T, and N. notoginsengisoli JCM 30743T. (A) Venn diagram was plotted using the OrthoVenn2 program to illustrate the distribution of orthologous clusters among these strains. The numbers in the overlapped regions of the Venn diagram represent the counts of CDS (coding DNA sequences) numbers that are shared by the genomes. (B) The number of orthologs in each genome used for generating the Venn diagram.
N. endophyticus BRMEA1T, obtained from the medicinal plant Selaginella involvens, has the potential to produce a variety of valuable secondary metabolites. With the use of WGS technology, biosynthetic gene clusters responsible for the synthesis of natural compounds in the entire genome of N. endophyticus BRMEA1T were predicted with antiSMASH. The results of genome mining are presented in Table 2. Four secondary metabolite biosynthesis gene clusters (SM-BCGs) were predicted from the entire genome of N. endophyticus BRMEA1T, including lasso peptides, linear azo(in)e-containing peptide, ribosomally synthesized and posttranslationally modified peptide (RiPP)-like terpenes, and type III polyketide synthase (T3PKS). Lasso peptides are a type of RiPPs found in bacteria such as Streptomyces (Arulprakasam and Dharumadurai, 2021; Kim and Kwak, 2021), Bacillus (Rahimi et al., 2018), and Thermogemmatispora (Komaki et al., 2016). These peptides have potential applications in various medical fields such as gastrointestinal disease, cardiovascular disease, Alzheimer's disease, tuberculosis, fungal infections, and cancer treatment; these are due to their antimicrobial, enzyme inhibition, receptor-blocking, anticancer, and human immunodeficiency virus antagonist properties (Bratovanov et al., 2020; Cheng and Hua, 2020; Knappe et al., 2008). Additionally, lasso peptides exhibit stability against heat treatment and extreme pH (Cheng and Hua, 2020), suggesting their potential role in the thermotolerance of N. endophyticus BRMEA1T.
The analysis of the gene cluster (Table 2, Supplementary Figs. 6–9) revealed that lasso peptide synthesis in N. endophyticus BRMEA1T involves three core biosynthetic genes: transglutaminase-like superfamily, glutamine amidotransfer-ase domain, and coenzyme PQQ synthesis protein D, along with an additional biosynthetic gene. Lasso peptides are natural products frequently identified in the phylum A ctino-mycetota, such as Streptomyces (Cheng and Hua, 2020), and include bacterially synthesized antimicrobial peptides that are assembled into ribosomes, belong to the class of posttranslationally modified peptides (RiPPs), and are characterized by cationic and amphipathic properties. This suggests that the antimicrobial activity of these bacteria may be linked to membrane permeabilization (Harwood et al., 2018). Lasso peptides exhibit diverse biological functions, including antimicrobial (Tenea and Ortega, 2021), antiviral, and antifungal activities that assist plants in defending against pathogen infection (Eyles et al., 2021). More than 80 lasso peptides have been characterized, including anantinB1, arcumycin, sviceucin, siamycin, chaxapeptin, steptomonomicin, and propeptin.
Terpenes have been gaining attention due to their various health benefits and medical applications. These natural compounds, found in essential oils, have demonstrated anticancer, antimicrobial, antiviral, antimalarial, antioxidant, and anti-inflammatory properties (De Sousa et al., 2015; de Souza Pedrosa et al., 2020; Kaur et al., 2019; Petrović et al., 2019; Stahl and Sies, 2005). Although there are no reports of terpenes contributing to bacterial heat tolerance, terpenes have been found to be one strategy of plants to enhance heat tolerance and alleviate abiotic/biotic stress (Hara 2020; Li et al., 2016; Schnitzler et al., 2010). For example, monoterpenes produced by plants have antibacterial, insecticidal, and anthelmintic properties, and have been found to enhance heat tolerance in plants through the expression of heat shock protein genes (Hara, 2020). T3PKSs are widely used in nutraceuticals and medicines, and microbial T3PKSs have significant biological functions and pharmaceutical activities, particularly those related to antimicrobial activity (Katsuyama and Ohnishi, 2012; Somashekaraiah et al., 2021). Phenolic lipids withered by T3PKS, such as ArsB in Azotobacter vinelandii, have been proposed to confer resistance to desiccation and heat in this bacterium (Funabashi et al., 2008). In conclusion, these valuable natural compounds seem to be perfectly suited to the characteristics of their respective host plants. This research highlights the potential use of thermotolerance genes and SM-BCGs to enhance stress tolerance and engineer crops resistant to various challenges. For example, previous studies explored bacterial strains producing pyomelanin, which aids grapes in preventing UV damage (Jiang et al., 2021), and nostoxanthin, known to reduce reactive oxygen species and improve salt stress (Jiang et al., 2023). These findings indicate that the BRMEA1T strain might play a significant role in facilitating thermotolerance adaptation in host plants.
In this study, our whole-genome-based comparative genomics analysis has shed light on bacterial adaptations to high-temperature environments. Our results revealed that thermotolerant bacteria possess a relatively high number of genes and proteins. Notably, N.endophyticus BRMEA1T was found to biosynthesize several secondary metabolites that are consistent with those of the host plant, which may con-tribute to its thermotolerance traits. These findings provide valuable genomic-based insights that could have potential applications in improving plant thermotolerance. Furthermore, our results suggest that secondary metabolites from bacterial strains may play a role in the regulation of thermotolerance in host plants, opening up new avenues for further search in this area.
The GenBank accession number for the whole-genome sequence of Neobacillus endophyticus BRMEA1T obtained in this study is JABRWH000000000. The NCBI BioProject and BioSample numbers are PRJNA634211 and SAMN14986670, respectively. The strain is accessible at the Korean Collection for Type Cultures (KCTC 43028T) and at the China Center for Type Culture Collection (CCTCC AB 2020071T).
Electronic Supplementary Material
Supplementary materials are available at Research in Plant Disease website (http://www.online-rpd.org/).
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This research was supported by the Korea Institute of Plan-ning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Agricultural Machinery/Equipment Localization Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321057051HD020), and by the KRIBB research ini-tiative program (KGM5282331).


