First Report of Pectobacterium versatile as the Causal Pathogen of Soft Rot in Kimchi Cabbage in Korea
Article information
Abstract
In September 2021, gray-to-brown discoloration and expanding water-soaked lesions were observed on the outer and inner layers and the core of kimchi cabbage (Brassica rapa subsp. pekinensis) in fields located in Samcheok, Gangwondo, Korea. A bacterial strain designated as KNUB-02-21 was isolated from infected cabbage samples. Phylogenetic analysis based on the sequences of the 16S rRNA region and the dnaX, leuS, and recA genes confirmed that the strain was affiliated with Pectobacterium versatile. Additionally, the biochemical and morphological profiles of the isolate were similar to those of P. versatile. Based on these results, the isolate was identified as a novel strain of P. versatile. Healthy kimchi cabbage slices developed soft rot upon inoculation with P. versatile KNUB-02-21 and exhibited symptoms similar to those observed in the diseased plants in fields. The re-isolated strains were similar to those of P. versatile. Prior to our study, P. versatile as the causative pathogen of kimchi cabbage soft rot had not been reported in Korea.
In Korea, kimchi cabbage (Brassica rapa subsp. pekinensis) is grown in the mountains in summer and in the lowlands during fall to avoid high temperatures and disease out-breaks. Kimchi cabbage had an average cultivation area of 12,722 ha from 2019 to 2021. In Gangwondo, kimchi cabbage was cultivated over an area of approximately 1,482 ha between 2020 and 2021 (Korean Statistical Information Service, 2022). Various diseases of cabbage have been reported worldwide. Verticillium dahliae, Fusarium equiseti, and Erysiphe cruciferarum are the main fungal pathogens causing stem decay diseases in Korea (Afroz et al., 2021; Dumin et al., 2021; Jee et al., 2008). In addition, soft rot disease caused by Pectobacterium species results in significant economic losses to dicotyledonous and monocotyledonous plants, as well as agricultural production businesses. Pectobacterium species produce various plant-cell-wall degrading enzymes, including cellulase, polygalacturonase, and pectinase, which are involved in causing soft rot in several plant hosts (Lee et al., 2014). Among several phytopathogenic Pectobacterium species, P. brasiliense and P. odoriferum have been reported to be causative pathogens of cabbage soft rot in Korea (Lee et al., 2014, 2021).
In September 2021, severe soft rot symptoms, such as gray-to-brown discoloration and expanding water-soaked lesions, were observed on kimchi cabbages in Samcheok, Gangwondo, Korea (Fig. 1). These symptoms are typical characteristics of bacterial diseases (bacterial soft rot) (Charkowski, 2018). Infected cabbage samples were used to isolate the causative pathogens. Small pieces of infected tissue were immersed in 5 ml of saline solution (0.8% NaCl) for 20 min. The resulting suspension was divided into 50 µl and spread on King's B (Difco, Sparks, MD, USA) and nutrient agar (Difco) media and incubated at 30°C for 48 hr. The cultures were plated twice using these media for purification. A bacterial strain, designated KNUB-02-21, was isolated from infected kimchi cabbages.

Soft rot disease caused by Pectobacterium versatile KNUB-02-21 in kimchi cabbage in the field at Samcheok, Gangwondo, Korea. (A) Symptoms of soft rot on the outer layer of kimchi cabbage. (B) Symptoms of soft rot on the inner layer of kimchi cabbage. (C) Symptoms of soft rot in the core of kimchi cabbage.
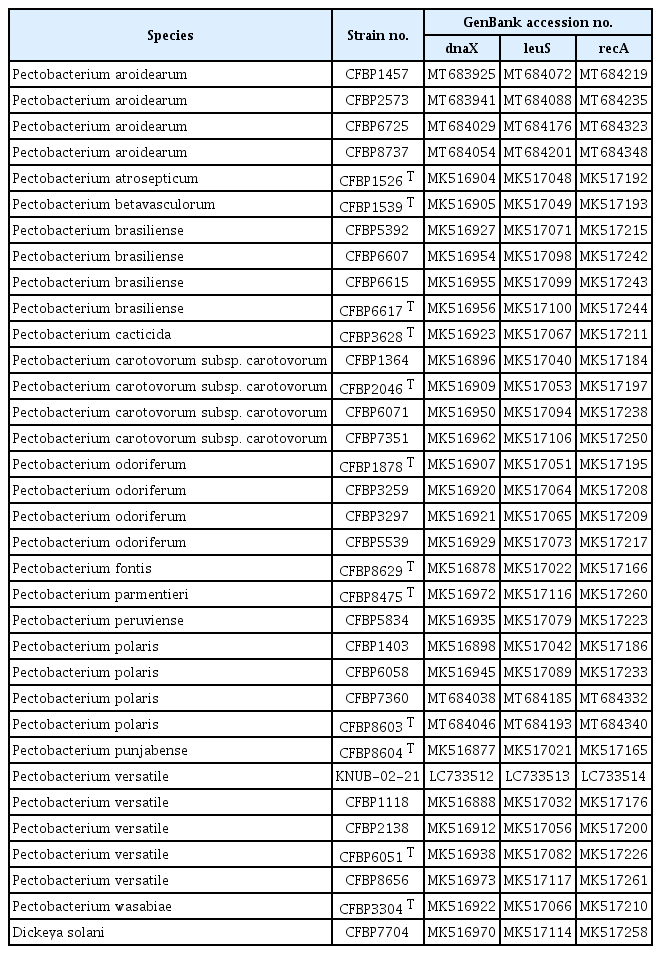
For molecular analysis, total genomic DNA was extracted from the bacterial culture after 24 hr of growth on nutrient agar medium using a commercial extraction kit (HiGene Genomic DNA Prep Kit, Biofact, Daejeon, Korea), following the manufacturer's instructions. The polymerase chain reaction (PCR) protocol and primers for the 16S rRNA region were adapted from Weisburg et al. (1991) (Supplementary Table 1). ExoSAP-IT PCR Product Cleaning Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used to purify the PCR products. The 16S rRNA region sequence of KNUB-02-21 (GenBank no. LC733515) was 1,387 bp in length. A BLAST search of the National Center for Biotechnology Information (NCBI) data-base revealed that the sequence of the 16S rRNA region of KNUB-02-21 is highly similar to that of P. carotovorum subsp. carotovorum JB56 (GenBank no. MT579566) (99.86%), P. brasiliense HNP201711 (GenBank no. MN393933) (99.78%), P. versatile 14A (GenBank no. CP034276) (99.78%), and P. odoriferum WQC37 (GenBank no. MH778625) (99.64%). These results unequivocally demonstrated that the comparative analysis based solely on the 16S rRNA region sequence could not accurately identify strain KNUB-02-21 at the species level. Based on the 16S rRNA region, only the taxonomic relationship at the genus level could be with other species; however, strain affiliation to Pectobacterium species and subspecies could not be precisely determined. Recent phylogenetic research using three concatenated housekeeping genes (dnaX, leuS, and recA) has allowed the assignment of 114 strains of the P. carotovorum complex to a few novel species (Portier et al., 2019). Therefore, dnaX, leuS, and recA of strain KNUB-02-21 were amplified, sequenced (Supplementary Table 1), and used for phylogenetic analysis.
A sequence of 511 bp for dnaX (GenBank no. LC733512), 522 bp for leuS (GenBank no. LC733513), and 714 bp for recA (GenBank no. LC733514) were obtained from KNUB-02-21. Based on the similarity of the dnaX gene sequence, P. versatile JB56A (GenBank no. MW930747) (100%), P. carotovorum subsp. carotovorum JB46 (GenBank no. MT632643) (100%), P. atrosepticum TEIC3211 (GenBank no. JN663800) (100%), P. polaris CFBP8603 T (GenBank no. MT684046) (97.15%), P. parvum YT22221 (GenBank no. CP046377) (97.06%), and P. actinidiae PRI-B17 (GenBank no. ON960282) (96.84%) were the closest phylogenetic relatives of the novel isolate. The leuS gene sequence of KNUB-02-21 exhibited 99.81% similarity to P. parvum YT22221 (GenBank no. CP102749) and P. versatile ECC15 (GenBank no. CP102749), 99.23% similarity to P. carotovorum RC5297 (GenBank no. CP045097), 98.85% similarity to P. odoriferum JK2.1 (GenBank no. CP034938), 97.71% similarity to P. polaris NIBIO1006 (GenBank no. CP017481), 97.13% similarity to P. quasiaquaticum A398-S21-F17 (GenBank no. CP065178), 97.13% similarity to P. aquaticum A212-S19-A16 (GenBank no. CP086253), and 96.74% similarity to P. brasiliense BC1 (GenBank no. CP092039). The closely related P. carotovorum subsp. carotovorum BTC2 (GenBank no. MF314822), P. versatile 14A (GenBank no. CP034276), P. odoriferum JK2.1 (GenBank no. CP034938), and P. aquaticum A212-S19-A16 (GenBank no. CP086253) shared 99.72%, 99.58%, 97.75%, and 96.35% identity, respectively, with the recA gene sequence of strain KNUB-02-21. Similar to the 16S rRNA region, these results indicated that the comparative analysis based on the sequence of only one of these molecular markers did not allow precise identification of the closest relatives.
Multilocus sequence analysis is currently a popular technique for confirming with a higher degree of certainty the phylogenetic relationships of species within a genus. The combined sequences of dnaX, leuS, and recA genes are effective for the identification and classification of strains affiliated with the genus Pectobacterium (Portier et al., 2019). Using the NCBI BLAST search, the sequences of the three molecular markers of strain KNUB-02-21 were compared with those of the reference Pectobacterium species, and close phylogenetic neighbors were selected (Table 1). The MEGA 7 software was used to conduct phylogenetic and molecular evolutionary analyses (Kumar et al., 2016). Maximum likelihood analysis was performed using Kimura's two-parameter model and the nearest neighbor interchange heuristic search method. Bootstrap analysis was performed with 1,000 replicates to assess the stability and validity of various clusters. In the phylogenetic tree, the novel isolate KNUB-02-21 and several strains of P. versatile clustered together in a monophyletic clade with a high bootstrap value, indicating their affiliation with the same species (Fig. 2).
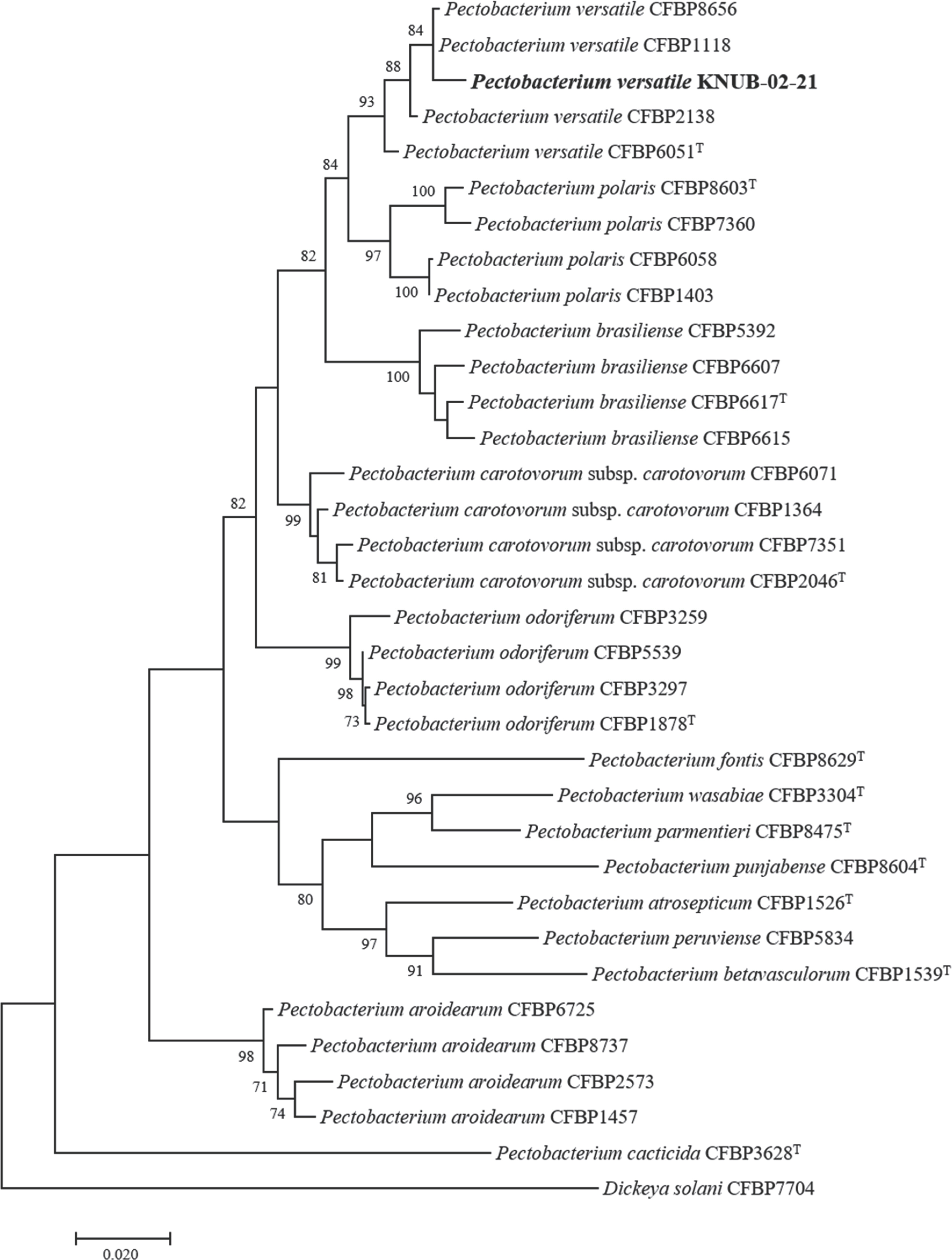
Maximum-likelihood phylogenetic tree, based on concatenated partial sequences of dnaX, leuS, and recA genes, depicting the phylogenetic position of strain KNUB-02-21 among related species of the genus Pectobacterium. Bootstrap values (based on 1,000 replications) greater than 70% are displayed on the branch points. Dickeya solani CFBP7704 was used as the outgroup. Scale bar=0.020 substitutions per nucleotide position.
According to Portier et al. (2019), a combination of three reactions, namely assimilation of D-arabitol, stachyose, and guanidine hydrochloride or L-galactonic acid-γ-lactone can help differentiate P. versatile, P. carotovorum subsp. carotovorum, P. odoriferum, P. brasiliense, P. actinidiae, P. aquaticum, and P. polaris. Isolate KNUB-02-21 responded positively and assimilated stachyose, L-galactonic acid-γ-lactone, and guanidine hydrochloride when tested using Biolog GEN III plates (Biolog, Hayward, CA, USA). However, the strain KNUB-02-21 tested negative for D-arabitol assimilation in the Biolog GEN III test. P. versatile exhibits a similar response towards the four growth substrates (Table 2), further confirming the affiliation of KNUB-02-21 with this Pectobacterium species. The results of the phylogenetic analysis and biochemical tests supported the identification of KNUB-02-21 as a novel strain belonging to P. versatile.
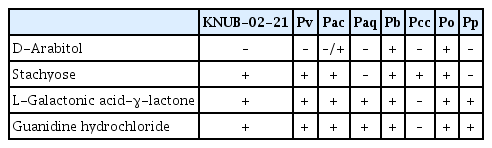
Biochemical test results of Pectobacterium versatile KNUB-02-21 and the most common soft rot pathogen Pectobacterium spp.
Pathogenicity tests were conducted on kimchi cabbage to confirm Koch's postulates. The surfaces of the kimchi cabbage slices were disinfected using 70% ethanol and washed with distilled water prior to inoculation. Three kimchi cabbage slices were pierced with a sterilized blue tip and inoculated with 20 µl of P. versatile KNUB-02-21 suspension at a concentration of 1×108 cells/ml. The plant used as an uninfected control was treated similarly but injected with 20 µl of distilled water. Slices of inoculated kimchi cabbage were maintained under greenhouse conditions (25–30°C, 80% relative humidity). After inoculation, the kimchi cabbage slices exhibited symptoms of soft rot after one day. After two days, the symptoms of gray-to-brown discoloration were similar to those observed previously, and increased water-soaked appeared on the kimchi cabbage (Fig. 3A, B, D, E). However, uninoculated kimchi cabbage slices did not develop any symptoms (Fig. 3C, F). Each slice of the infected kimchi cabbage was used to re-isolate the pathogen. In each case, the isolated bacterial strain was re-identified as P. versatile (data not shown).

Effects of Pectobacterium versatile KNUB-02-21 inoculation on kimchi cabbage slices. (A, B, D, E) Development of gray-to-brown discoloration and expanding water-soaked lesions in infected specimens after two days, similar to the soft rot disease symptoms observed first in kimchi cabbage. (C, F) After two days, no symptoms were observed in distilled water-inoculated, uninfected kimchi cabbage slices.
The host range of P. versatile KNUB-02-21 was tested using four Brassicaceae plant species: beta (Beta vulgaris subsp. vulgaris), bok choy (Brassica rapa subsp. chinensis), lettuce (Lactuca sativa), and kale (Brassica oleracea). P. versatile KNUB-02-21 was suspended in sterile water, and the bacterial concentration was adjusted to 1×108 cells/ml based on spectro-photometric readings (absorbance at 600 nm). Leaves from harvested plants were pierced with a sterilized blue tip and inoculated by injecting 20 μl of bacterial suspension into the petiole and midrib of each leaf. The plant used as the uninfected control sample was injected with distilled water. All the plants were maintained under greenhouse conditions (25–30°C, 80% relative humidity). After bacterial inoculation, the four Brassicaceae plant species presented symptoms of soft rot after two days (Table 3, Fig. 4).

Host range test for Pectobacterium versatile KNUB-02-21. Plants were inoculated with bacterial cells at 1×108 cells/ml, and incubated for two days in the greenhouse: beta (A), bok choy (B), lettuce (C), kale (D), and uninoculated Brassicaceae plants (E–H), respectively. The arrowheads indicate the area where the inoculation was performed.
Several species of the genus Pectobacterium cause disease in various host plants. P. versatile has been identified as the causative agent of blackleg and soft rot in various plants (Chen et al., 2022; Ma et al., 2021). P. versatile has been isolated from artichokes, bittersweet, cabbage, carrot, chicory, chrysanthemum, cyclamen, hyacinth, iris, leek, potato, and primrose (Portier et al., 2020). The phytopathogenic species P. versatile has been reported in Algeria, Canada, Finland, France, Morocco, the Netherlands, Spain, Syria, the United Kingdom, and the United States (Portier et al., 2020).
In this study, P. versatile was identified for the first time in Korea as a pathogen of soft rot in kimchi cabbage. In addition, this Pectobacterium species was found to cause a similar soft rot disease in four other plants, namely, beta, bok choy, lettuce, kale and cabbage of the family Brassicaceae. Our results expand our understanding of P. versatile distribution in Korea, enhance our knowledge of the causative pathogens of soft rot in kimchi cabbage, and can be applied to improve control strategies to prevent economic losses.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This study was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ017000)” Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at Research in Plant Disease website (http://www.online-rpd.org/).
Supplementary Table 1.
Polymerase chain reaction (PCR) protocol and primers used to amplify the 16S rRNA region and three housekeeping genes (dnaX, leuS, and recA) in this study