Ahmadpour, S. A., Farokhinejad, R. and Mehrabi-Koushki, M. 2017. Further characterization and pathogenicity of
Didymella microchlamydospora causing stem necrosis of
Morus nigra in Iran.
Mycosphere 8: 835-852.

Aveskamp, M. M., de Gruyter, J., Woudenberg, J. H. C., Verkley, G. J. M. and Crous, P. W. 2010. Highlights of the
Didymellaceae: a poly-phasic approach to characterise
Phoma and related pleospora-lean genera.
Stud. Mycol. 65: 1-60.



Aveskamp, M. M., Verkley, G. J. M., De Gruyter, J., Murace, M. A., Perelló, A., Woudenberg, J. H. C. et al. 2009. DNA phylogeny reveals polyphyly of
Phoma section
Peyronellaea and multiple taxonomic novelties.
Mycologia 101: 363-382.


Barilli, E., Cobos, M. J. and Rubiales, D. 2016. Clarification on host range of
Didymella pinodes the causal agent of pea ascochyta blight.
Front. Plant Sci. 7: 592.



Bewket, W. 2009. Rainfall variability and crop production in Ethiopia case study in the Amhara region. In: Proceedings of the 16th International Conference of Ethiopian Studies, eds. by S. Ege, H. Aspen, B. Teferra and S. Bekele, pp. 823-836. Norwegian University of Science and Technology, Trondheim, Norway.
Chen, Q., Hou, L. W., Duan, W. J., Crous, P. W. and Cai, L. 2017.
Didymellaceae revisited.
Stud. Mycol. 87: 105-159.



Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L. and Crous, P. W. 2015. Resolving the Phoma enigma. Stud. Mycol. 82: 137-217.
Chungu, D., Muimba-Kankolongo, A., Wingfield, M. J. and Roux, J. 2010. Identification of fungal pathogens occurring in eucalyptus and pine plantations in Zambia by comparing DNA sequences.
Forestry 83: 507-515.

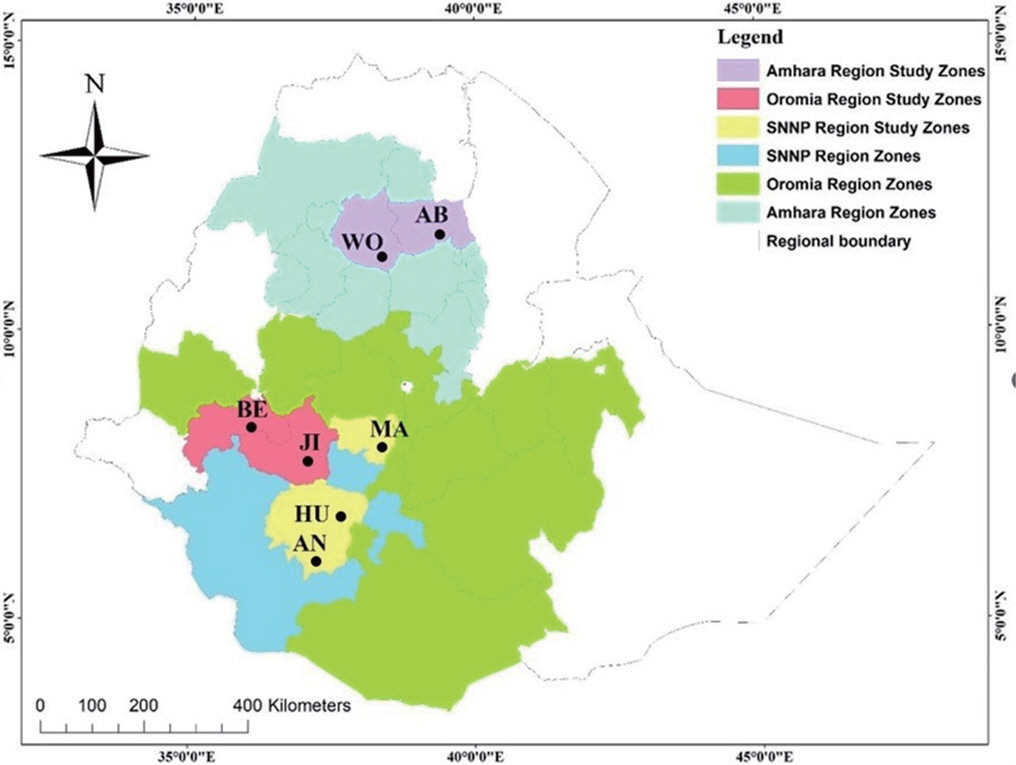
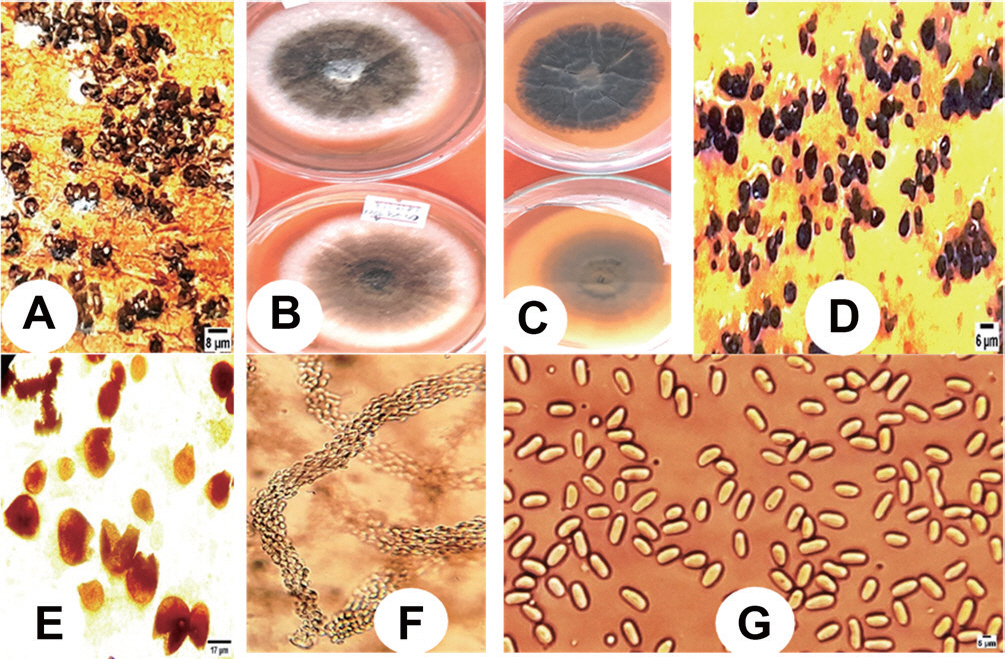
Darge, W. A. 2017. Diversity of pathogenic fungi on plantation forests of North and North-West Ethiopia.
Int. J. Phytopathol. 6: 27-34.


Darge, W. A. and Bogale, A. T. 2017.
Eucalyptus globulus (
E. globulus) leaf spot and stem canker diseases due to
Phoma spp. in North and North-West Ethiopia.
Int. J. Plant Pathol. 8: 14-22.

Demissie, A. G., Darge, W. A. and Cafà, G. 2020.
Neofusicoccum parvum causing
Eucalyptus canker and dieback diseases in Ethiopia.
Int. J. Plant Pathol. 11: 1-5.

Dessie, A. B., Abtew, A. A. and Koye, A. D. 2019. Determinants of the production and commercial values of
Eucalyptus woodlot products in Wogera District, Northern Ethiopia.
Environ. Syst. Res. 8: 4.


Dessie, G. and Erkossa, T. 2011.
Eucalyptus in East Africa, Socio-economic and Environmental Issues. Planted Forests and Trees Working Paper FP46/E. Food and Agriculture Organization, Rome, Italy. 30 pp.
El-Gali, Z. I. and El-Zahaf, B. S. 2015. Status and symptomatology of Alternaria alternata ceratoni blight of carob (Ceratoni asiliqua L.) in adjoining areas of El-Beida City-Libya. Sky J. Microbiol. Res. 3: 030-035.
Food and Agriculture Organization. 2009.
Eucalyptus in East Africa, the Socio-economic and Environmental Issues. Food and Agriculture Organization, Addis Ababa, Ethiopia. 30 pp.
Food and Agriculture Organization. 2020. Global forest resources assessment: 2020 main report. Food and Agriculture Organization, Rome, Italy. 184 pp.
Gebremichael, A., Quraishi, S. and Mamo, G. 2014. Analysis of seasonal rainfall variability for agricultural water resource management in southern region, Ethiopia. J. Nat. Sci. Res. 4: 56-79.
Gezahgne, A., Cortinas, M.-N., Wingfield, M. J. and Roux, J. 2005. Characterization of the Coniothyrium stem canker pathogen on Eucalyptus camaldulensis in Ethiopia. Australas. Plant Pathol. 34: 85-90.
Gezahgne, A., Roux, J., Hunter, G. C. and Wingfield, M. J. 2006.
Mycosphaerella species associated with leaf disease of
Eucalyptus globulus in Ethiopia.
For. Pathol. 36: 253-263.

Gezahgne, A., Roux, J. and Wingfield, M. J. 2003. Diseases of exotic Eucalyptus and Pinus species in Ethiopia plantations. S. Afr. J. Sci. 99: 29-33.
Golani, M., Abbo, S., Sheman, A., Frenkel, O. and Shtienbrg, D. 2016. The temperature response and aggressiveness of
Peyronellaea pinodes isolates originating from wild and domesticated
Pisum sp. in Israel.
Phytopathology 106: 824-832.


Hall, T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95-98.
Keirnan, E. C., Tan, Y. P., Laurence, M. H., Mertin, A. A., Liew, E. C. Y., Summerell, B. A. et al. 2021. Cryptic diversity found in
Didymellaceae from Australian native legumes.
MycoKeys 78: 1-20.




Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA 10: Molecular Evolutionary Genetics Analysis across computing platforms.
Mol. Biol. Evol. 35: 1547-1549.


Lemenih, M. and Kassa, H. 2014. Re-greening Ethiopia: history, challenges and lessons.
Forests 5: 1896-1909.

Li, G. Q., Liu, F. F., Li, J. Q. and Chen, S. F. 2018.
Botryosphaeriaceae from
Eucalyptus plantations and adjacent plants in China.
Persoonia 40: 63-95.



Li, G., Slippers, B., Wingfield, M. J. and Chen, S. 2020. Variation in
Botryosphaeriaceae from
Eucalyptus plantations in YunNan province in southwestern China across a climatic gradient.
IMA Fungus 11: 22.




Maid, M. and Ratnama, W. 2014. Incidences and severity of vascular wilt in
Acacia mangium plantations in Sabah, Malaysia.
AIP Conf. Proc. 1614: 784-789.

Masood, A., Saeed, S., Iqbal, N., Malik, M. T. and Kazmi, M. R. 2010. Methodology for the evaluation of symptoms severity of mango sudden death syndrome in Pakistan. Pak. J. Bot. 42: 1289-1299.
Mesfin, A. and Wubalem, T. 2014.
Eucalyptus in Ethiopia: risk or opportunity?”. URL
http://www.eiar.gov.et [1 August 2022]
Mohali, S. R., Slippers, B. and Wingfield, M. J. 2007. Identification of Botryosphaeriaceae from Eucalyptus, Acacia and Pinus in Venezuela. Fungal Divers. 25: 103-125.
Noordeloos, M. E., De Gruyter, J., Van Eijk, G. W. and Roeijmans, H. J. 1953. Production of dendritic crystals in pure cultures of
Phoma and
Ascochyta and its value as a taxonomic character relative to morphology, pathology and cultural characteristics.
Mycol. Res. 97: 1343-1350.

Old, K. M., Wingfield, M. J. and Yuan, Z. Q. 2003. A Manual of Diseases of Eucalyptus in South-East Asia. Center for International Forestry Research, Bogor, Indonesia. 104 pp.
Pérez, C. A., Wingfield, M. J., Slippers, B., Altier, N. A. and Blanchette, R. A. 2009.
Neofusicoccum Eucalyptorum, Eucalyptus pathogen, on native Myrtaceae in Uruguay. Plant Pathol. 58: 964-970.
Pillay, K., Slippers, B., Wingfield, M. J. and Gryzenhout, M. 2013. Diversity and distribution of co-infecting
Botryosphaeriaceae from
Eucalyptus grandis and
Syzygium cordatum in South Africa.
S. Afr. J. Bot. 84: 38-43.

Sánchez Márquez, S., Bills, G. F. and Zabalgogeazcoa, I. 2010. Fungal species diversity in juvenile and adult leaves of
Eucalyptus globulus from plantations affected by Mycosphaerella leaf disease.
Ann. Appl. Biol. 158: 177-187.

SAS Institute Inc. 2011. SAS 9.3 system options: reference. 2nd ed. SAS Institute Inc., Cary, NC, USA.
Slippers, B. and Wingfield, M. J. 2007.
Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact.
Fungal Biol. Rev. 21: 90-106.

Silva, X., Roux, J. and Asiegbu, F. O. 2020. Diseases of
Eucalypts in Paraguay and first report of
Theratosphaeria zuluensis from South America.
Forests 11: 1035.

Tadesse, W., Gezahgne, A., Tesema, T., Shibabaw, B., Tefera, B. and Kassa, H. 2019. Plantation forests in Amhara region: challenges and best measures for future improvements. World J. Agric. Res. 7: 149-157.
Tadesse, W., Gezahgne, A., Tesema, T., Shibabaw, B., Tefera, B. and Kassa, H. 2019. Plantation forests in Amhara region: challenges and best measures for future improvements. World J. Agric. Res. 7: 149-157.
Tamura, K. 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases.
Mol. Biol. Evol. 9: 678-687.

Tamura, K., Nei, M. and Kumar, S. 2004. Prospects for inferring very large phylogenies by using the neighbour-joining method.
Proc. Natl. Acad. Sci. U. S. A. 101: 11030-11035.


Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eb-erhard, U. et al. 2019. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation.
Stud. Mycol. 92: 135-154.



White, T. J., Bruns, T., Lee, S. and Taylot, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications, eds. by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, pp. 315-322. Academic Press, San Diego, CA, USA.

Wingfield, M. J., Slippers, B., Hurley, B. P., Coutinho, T. A., Wingfield, B. D. and Roux, J. 2008. Eucalypt pests and diseases: growing threats to plantation productivity.
South. For. 70: 139-144.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print