Morphology and Molecular Characteristics of Alternaria sonchi Causing Brown Leaf Spot on Sonchus asper in Korea
Article information
Abstract
During a disease survey on weeds and minor cultivated crops in Korea, a brown leaf spot disease was observed on Sonchus asper. Leaf lesions were round or irregular in shape, and grayish brown to brown with a purple margin. In severe infection, lesions enlarged and coalesced, resulting in blighting of the leaves. The isolates from these leaf lesions were identified as Alternaira sonchi based on morphological characteristics and phylogenetic analyses of Internal transcribed spacer region, Alternaria allergen a1, glyceraldehyde 3-phosphate dehydrogenase, RNA polymerase II, and translation elongation factor genes. This study provides a comprehensive description of the morphological characteristics and phylogenetical traits of A. sonchi causing brown leaf spot on S. asper in Korea.
Introduction
Sonchus asper (L.) Hill is an annual or biennial plant in the dandelion tribe within the Asteraceae (Moerman, 1998). It is considered a noxious, invasive weed in many regions (Akhtar, 2019; Auld et al., 1987), but its tender young leaves and stem tops can be used as a vegetable and added to salads (Facciola, 1990; Tanaka, 1976), and its peeled stems can be eaten raw (Moerman, 1998). S. asper is sometimes cultivated locally as a food plant in New Zealand (Brooker et al., 1989); in other countries it is grown as a medicinal plant for its antibacterial effects (Khan et al., 2010; Upadhyay et al., 2013). In Korea, the plant is found in cultivated soil, pastures, orchards, roadsides, edges of yards, waste areas and in grasslands.
The genus Alternaria was originally characterized by production of dark-colored phaeodictyospores singly or in chains. Almost 275 Alternaria species have been described based upon their morphological characteristics (Simmons, 2007), and the phylogenetic relationships within Alternaria and their allied genera have been delineated based on nucleotide sequence data of the 18S nuclear ribosomal DNA (nrDNA), 28S nrDNA, internal transcribed spacer (ITS) region, and regions of the gyceraldehyde-3-phosphate dehydrogenase (GAPDH), RNA polymerase II (RPB2), and translation elongation factor (TEF1)-alpha genes (Woudenberg et al., 2013). To date, A. sonchi, A. simmonsii, and A. alternata have been reported on leaves of Sonchus spp. including S. asper, S. arvensis, and S. oleraceus from several countries in Asia, Europe, Africa, and North America (Akhtar, 2019; Ellis, 1971; Simmons, 2007; Zhang et al., 2021). In Korea, no specifically related information recorded for genus Alternaria causing diseases on Sonchus spp. (National Institute of Agricultural Sciences and The Korea Society of Plant Pathology, 2021).
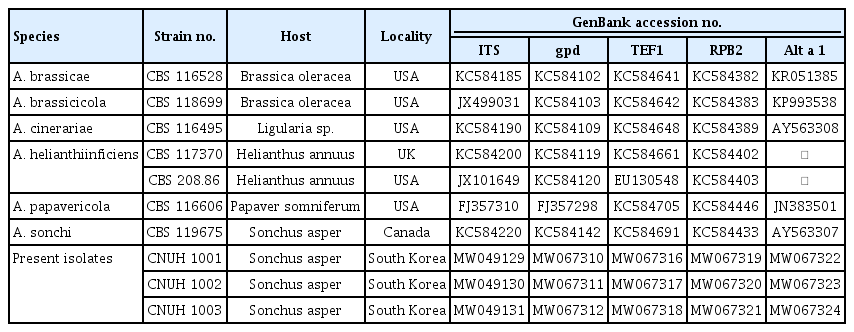
During the course of disease survey on weeds and minor cultivated crops in Korea in September 2019, a brown leaf spot disease of S. asper was observed in gardens and roadsides in Gongju and Nonsan, Chungnam Province, Korea. Leaf spots were round or irregular in shape, variable in size, grayish brown, or brown with a purple margin (Fig. 1A). The spots often enlarged and coalesced, resulting in blighting of the leaves (Fig. 1B), which is corresponded to the symptoms caused by genus Alternaria. In severe infection, incidence rate of the disease was more than 40%.
Symptoms of leaf spot on Sonchus asper caused by Alternaria sonchi. (A, B) Field symptoms: early symptoms of the disease (A) and severely infected symptoms of the disease (B). (C-E) Pathogenicity assay on 2-month-old Sonchus asper plant using A. sonchi suspension (1×105 spores/ml): initial symptoms on leaves post-24 hr-inoculation (C) and brown spots on plant petiole post-3 day-inoculation (D), and brown spots symptoms on leaves post-5 day-inoculation (E).
The objective of the present study was to describe this brown leaf spot disease of S. asper and to identify the causal agent based on the morphological characteristics and phylogenetic analyses of five genes (ITS, Alternaria allergen a 1 [Alt a1], GAPDH, RPB2, and TEF1). We also aimed to provide a detailed morphological description of A. sonchi for future taxonomic study.
Materials and Methods
Isolation of fungus. Six symptomatic plant samples of Sonchus asper were collected during fall 2019 in Chungnam Province and examined microscopically for the presence of Alternaria at the day of collection or after incubation on well moistened blotters in Petri dish at 23°C in a 16 hr dark/8 hr near-ultraviolet light regime. Isolates of Alternaria were obtained by single spore isolation method using a sterile glass needle under stereomicroscope (SOPTOP MP50, China), and cultured on potato dextrose agar (PDA; Difco, Sparks, MD, USA). A total 12 isolates of Alternaria were obtained and three isolates (CNUH 1001, CNUH 1002, CNUH 1003) were randomly selected to assess morphological and phylogenetic characteristics. The isolates were stored in 20% glycerol at -80°C at the Culture Collection of Chungnam National University.
Morphological observations. The isolate CNUH 1001 was used for morphological description. The isolate was point inoculated on potato carrot agar (PCA; 20 g white potato, 20 g carrot, and 20 g agar in 1 l distilled water) and V-8 juice agar (200 ml V8 juice, 3 g CaCO₃, 20 g agar in 1 l distilled water) and incubated for 9 days at 22-23°C under a fluorescent light/dark cycle of 8/16 hr, as previously recommended (Simmons and Roberts, 1993). Conidiophores and conidia from incubated leaf pieces on well moistened blotters and from the surface of 10-day-old cultures were mounted in plain lactophenol and covered with a cover glass. Morphological characteristics such as conidial shape, size, color, surface ornamentation and number of septa were observed under a compound microscope (Olympus BX50, Tokyo, Japan) with a DP21 digital camera (Olympus) at 400× magnification. Measurements of conidia and conidiophores were made from at least 80 replicates of all relevant morphological features. For producing macroscopic characters such as growth and color, pure cultures of the isolates were grown on PDA, PCA and V-8 juice agar medium at 23°C, darkness, 7 days.
DNA extraction and PCR amplification. For phylogenetic analysis, the three isolates (CNUH 1001, CNUH 1002, and CNUH 1003) were grown on PDA for 7 days at 23°C. Mycelia were collected from the growing colonies on PDA plates for DNA extraction. Total DNA was extracted from growing fresh hyphae using Quick-DNA Fungal Miniprep Kit (Zymo Research, Irvine, CA, USA). The DNA pellet was stored at -20°C for further use. DNA samples were visualized under ultraviolet illumination by gel electrophoresis, amounts and qualities were compared with standard markers. The genes used to characterize the isolates were: ITS region containing ITS-5.8S-ITS2 of nrDNA operon amplified with ITS5 and ITS4 primers (White et al., 1990), the conserved region of Alt a1 gene amplified with primers Alt-a1-for and Alt-a1-rev (Hong et al., 2005), a portion of GAPDH amplified with primers gpd1 and gpd2 (Berbee et al., 1999), RPB2 amplified with primers RPB2-5F2 (Sung et al., 2007) and fRPB-7cR (Liu et al., 1999), and the TEF1-alpha gene amplified with primers EF1-728F and EF1-986R (Carbone and Kohn, 1999) or EF2 (O’Donnell et al., 1998). PCR amplification was performed in 20 μl (10 μl AccuPower TaqPCR PreMix, 7 μl ddH2O, 1 μl DNA, 1 μl of each primer), following the procedure described by Woudenberg et al. (2013). The PCR products were observed on 1% agarose gels. Successfully amplified DNA was purified using Gel and PCR Clean-up Kit (Cosmo Genetech, Seoul, Korea) and sequenced with the same primers.
Phylogenetic analyses. Sequences of all isolates were assembled and proofread using PHYDIT 3.2 software (Chun, 1995) and deposited in GenBank (Table 1). Each sequence from the strains was blasted for comparison on the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/) using BLASTn. The most highly matching sequences were downloaded, along with sequences from seven type species included for phylogenetic reference (Woudenberg et al., 2013) (Table 1). Multiple alignments were performed using the default settings of the MEGA 7.0 software (Kumar et al., 1994). Maximum likelihood (ML) analysis was carried out in MEGA 7.0 employing the best-fit model of molecular evolution with 1,000 bootstrap (BS) replicates were computed and BS values above 80% were considered highly supported.
Pathogenicity tests. Pathogenicity tests were carried out in the greenhouse on 2-month-old healthy plants of Sonchus asper. We sprayed three test plants with spore suspension (1×105 conidia/ml) of the isolate CNUH 1001. Plants inoculated with sterile distilled water were used as controls. After inoculation, plants were covered with polyethylene bags for 2 days at room temperature. Then the bags were removed, and plants were maintained at 24°C under a fluorescent light/dark cycle of 16/8 hr with 50% relative humidity. Disease progress was monitored for 7 days. Morphological and phylogenetical characteristics of the fungus re-isolated from symptomatic tissues of stems and leaves was compared with the original isolate to full with Koch’s postulates. Tests were repeated three times.
Results
Morphological observations. The cultural and morphological characteristics of the three present isolates CNUH 1001, CNUH 1002, and CNUH 1003 were consistent. The morphology characteristics of conidia and apical secondary conidiophores of the species summarized and compared with representative isolate A. sonchi E.G.S. 43.131 (=CBS 119675) in Table 2.
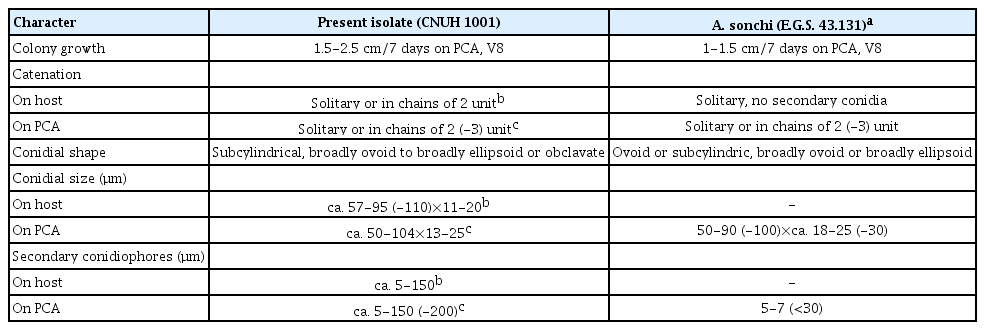
Comparison of morphological characteristics between present isolates and Alternaria sonchi representative isolate
Colonies on PDA, amphigenous, slow, stunted growth, reaching about 1.5-1.7 cm in diameter after 7 days (2.0–2.5 cm on both PCA and V8 agar media). Obverse cottony with abundant development of aerial mycelium, pale pinkish buff, reverse pale orange to orange, poor sporulation, scarifying the surface mycelium induces increased sporulation. Conidiophores (Fig. 2) arising singly, sometimes in groups of two or more, simple, occasionally branched, straight to slightly flexuous, commonly with a single terminal conidiogenous site, rarely proliferating sympodially and geniculate with 1 (-2) lateral conidiogenous site, pale brown, almost hyaline at the apex, smooth-walled, multi-septate, ca. 5-150 μm on host, ca. 5-150 (-200) μm on PCA and V8 agar. Sporulation pattern (Fig. 2-C) solitary or in short chains of 2 (-3) units through the agency of a secondary conidiophore (pseudorostrum), on agar media chains of 2-3 occurring more frequently than in nature, straight to slightly curved, long ellipsoid or subcylindric with blunt, rounded end cells. Conidia (Fig. 2D, E) first formed usually larger and nearly cylindrical to narrowly ovoid, subsequent secondary conidia becoming progressively smaller, pale to golden brown or olivaceous brown in color, smooth or minutely verruculose when juvenile, becoming punctate and verrucose with age, often slightly or distinctly constricted at the transverse septa of the conidium body, beakless, some have a short broadly tapered apical cell, some others have an apical secondary conidiophore analogous to the beak; conidium body ca. 57-95 (-110)×11-20 μm on host, with 4-7 transverse septa and occasionally 1-2 longitudinal septa, ca. 50-104×13-25 μm on PCA, with 4-7 transverse septa and occasionally 1-2 longitudinal septa, ca. 48-99×13-23 μm on V-8 juice agar, with 4-7 transverse septa and occasionally 1-2 longitudinal septa; secondary conidiophores simple and unbranched, cylindrical, septate, slightly lighter in color than the body, lightly swollen at the apex, initiating a secondary conidium, variable in length, some are short and 1-celled, ca. 5-65×4- 6 μm, many others are relatively long and septate, 100- 170 μm long, 5-7 μm wide, on host the apical secondary conidiophores often shorter than those on agar media (Table 2), occasionally a lateral secondary conidiophore produced from an intercalary conidium cell.
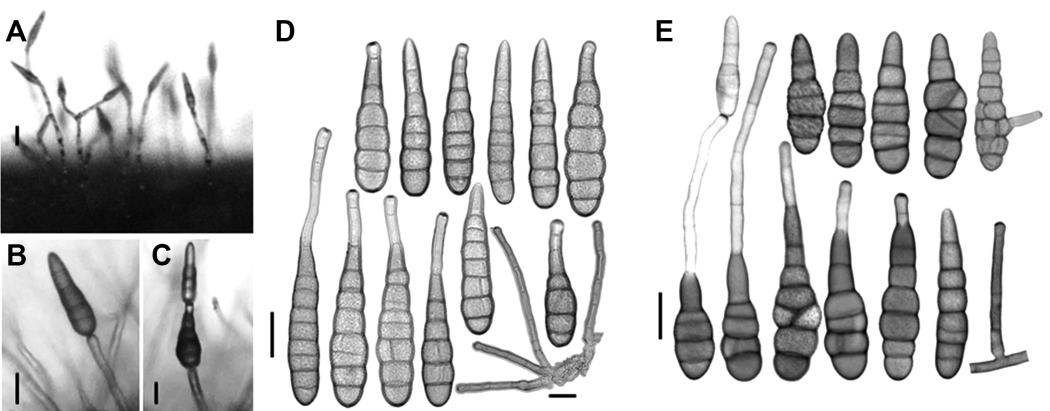
Morphology characteristics of Alternaria sonchi (CNUH 1001). (A-C) Sporulation pattern of Alternaria sonchi (CNUH 1001): from diseased leaves after 2 days of incubation at 23°C, 8 hr light (A) and from V8 medium after 9 days, 23°C, 8 hr light (B, C). (D) Conidia and conidiophores of Alternaria sonchi from diseased leaves after 2 days of incubation at 23°C, 8 hr light. (E) Conidia and conidiophores of Alternaria sonchi (CNUH 1001) from potato carrot agar medium after 9 days, 23°C, 8 hr light. Scale bars=25 µm.
Phylogenetic analyses. Gene sequence of ITS, Alt a1, GAPDH, RPB2, and TEF1 of the three isolates (CNUH 1001, CNUH 1002, and CNUH 1003) resulted in 589 bp, 470 bp, 580 bp, 794 bp, and 240 bp long, respectively. The sequences of all five genes were 100% identical among the three isolates. All resulting sequences were deposited in GenBank, accession numbers are shown in Table 1. The combined datasets of ITS, Alt a1, GAPDH, RPB2, and TEF1 resulted in an alignment containing a total of 2,484 characters including alignment gaps, which comprised 527, 391, 580, 742, and 244 characters for each of the five genes, respectively. The sequences of ITS, RPB2, and TEF1 gene were identical to those of the representative isolate (CBS 119675) of A. sonchi from Sonchus plants. One and four nucleotides of our isolates were different from CBS 119675 based on GAPDH and Alt a1 sequences, respectively (Supplementary Fig. 1A, B). Based on the combined phylogenetic tree, the three isolates had identical sequences for each of the five genes and formed a strongly supported clade (100% BS value) with the representative isolate (CBS 119675) of A. sonchi (Fig. 3).
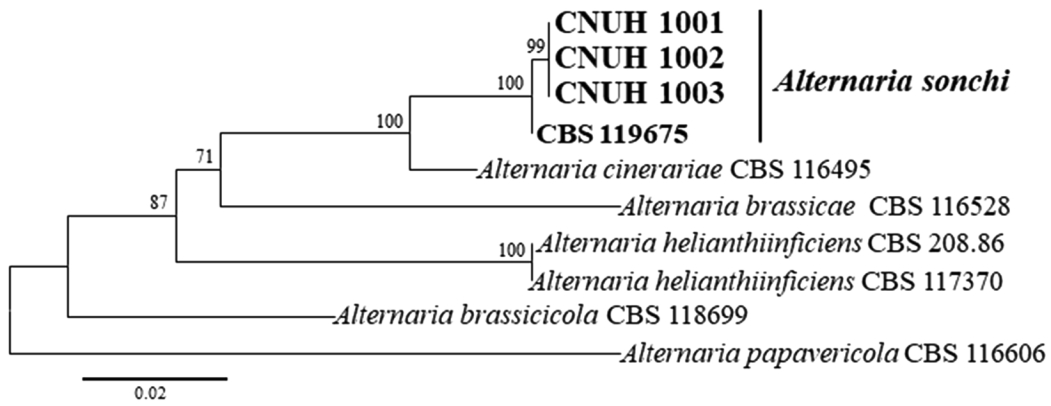
Maximum likelihood tree inferred from analysis of combined internal transcribed spacer, Alternaria allergen a 1, gyceraldehyde3-phosphate dehydrogenase, RNA polymerase II, and translation elongation factor gene sequences of Alternaria isolates from Sonchus and relevant species. Bootstrap values (≥70%) based on 1,000 replicates are shown at the nodes.
Pathogenicity tests. Initial symptoms on leaves of S. asper appeared 2-3 days after inoculation as small grayish brown spots, changing to pale yellowish-brown (Fig. 1C, D). Brown stripes were clearly observed on stems at 5 days after inoculation (Fig. 1E). All the symptoms resembled to those originally observed in the field. The same fungus was re-isolated from both leaves and stems and identified as the original inoculated fungus determined by morphological characteristics and phylogenetic analysis. Control plants did not show any symptoms. The results in agreement with Koch’s postulates and indicate that the fungus is the causal agent of the brown leaf spot disease in S. asper.
Discussion
The fungus we isolated from the leaf spots of Sonchus asper was identified as Alternaria sonchi J. J. Davis based on morphological and cultural characteristics, phylogenetic assessment and symptoms on S. asper.
According to conidial morphology, the fungus in this study have a subcylindrical, broadly ovoid or broadly ellipsoid spore body with a short broadly tapered apical cell or with a simple secondary conidiophore. These characters of the fungus are in general agreement with those of A. sonchi (Simmons, 1997, 2007; Woudenberg et al., 2013) (Table 2). However, our isolates a little bit differ from the representative isolate of A. sonchi (E.G.S. 35.193) in nature of the secondary conidiophores; our isolates produce both short and long secondary conidiophores on agar media and on natural substrata, but the representative isolate produces short secondary conidiophores only in culture (Simmons, 1997, 2007). According to Simmons (1997, 2007), secondary conidiophores involved in chain production of A. sonchi usually are a single cell ca. 5-10×5-7 μm and they seldom are longer than ca. 30 μm. While isolates in our study are variable in length; some are short, but many others are long (150-200 μm) and septate. Additionally, the representative isolate commonly forms short chains of 2 (-3) conidia only in culture, not in any field specimen (Simmons, 2007); however, our isolates readily forms short chains of 2 (-3) conidia not only in culture on agar media but also on natural substrata, although chains occurring more frequently in culture than in nature.
It is well known that conidial morphology of Alternaria spp. is variable based on culture conditions and substrate (Deng et al., 2015; Yu et al., 2003). In this investigation, the morphology of conidia and apical secondary conidiophores of A. sonchi showed variation according to culture substrate. The conidia formed on lesions of the host usually have narrower conidia and shorter secondary conidiophores than those produced on agar media (Table 2).
Sequence analyses of different types of genes such as ITS, Alt a1, ATPase, β-tubulin, endo-PG, GAPDH, RPB2, and TEF1 have been employed to classify or segregate Alternaria species in recent studies (Deng et al., 2013, 2015; Hong et al., 2005; Lawrence et al., 2013; Park et al., 2005; Peever et al., 2004; Pryor and Bigelow, 2003; Woudenberg et al., 2013; Yu et al., 2003). In the present study, the sequences of the three isolates were found to be 100 % identical to those of A. sonchi reference isolate (CBS 119675) based on the ITS, RPB2, and TEF1 region. In addition, there was no nucleotide difference among CBS 119675 and our isolates based on the ATPase gene sequence (data not shown). The three isolates were 100% clustered as a sister group to A. sonchi CBS 119675, according to the ML tree (Fig. 3) generated from a combined dataset of the ITS, Alt a1, GAPDH, RPB2, and TEF1 sequence data. Woudenberg et al. (2013) delineated phylogenetic lineages within Alternaria and allied genera based on nucleotide sequence data of parts of the 18S nrDNA, 28S nrDNA, ITS, GAPDH, RPB2, and TEF1-alpha gene regions (Woudenberg et al., 2013). They found that the Alternaria clade contained 24 internal clades and six monotypic lineages, and they treated the 24 internal clades as sections. A. sonchi associated with Sonchus plants belong to the section Sonchi along with A. cinerariae (Woudenberg et al., 2013). So far, accessible sequences data of A. sonchi from NCBI database were limited, our isolates’ data increased the variability for taxonomy study. The phylogenetic analyses along with the morphological characteristics indicates that the fungus isolated from S. asper is A. sonchi.
S. asper is well-known worldwide as a noxious weed with pappus-type seeds, that are wind dispersed over long distances, invading open and disturbed areas in a wide variety of environments such as cultivated fields, gardens, and roadsides (Upadhyay et al., 2013). A total of 29 fungal species are recorded to have caused different diseases on S. asper in the United States Department of Agriculture (USDA) fungus database (https://nt.ars-grin.gov/fungaldatabases/fungushost/fungushost.cfm); among these, Oidium sp., and Septoria sonchi have been reported in Korea (National Institute of Agricultural Sciences and The Korea Society of Plant Pathology, 2021). Powdery mildews caused by Golovinomyces cichoracearum and Leveillula taurica and leaf spot disease by Alternaria sonchi have been reported from S. asper in many countries (Elliott, 1916; Gautam and Avasthi, 2018; Park et al., 2008; Pei et al., 2012; Simmons, 2007). Leaf spot diseases on S. asper caused by A. alternata also reported recently in Pakistan (Akhtar, 2019) and China (Zhang et al., 2021). To our knowledge, our study first gave a comprehensive description of the morphological and phylogenic characteristics of A. sonchi, which causes brown leaf spots on S. asper in Korea.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This project was support by the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ014507022021)” Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at Research in Plant Disease website (http://www.online-rpd.org/).
Comparison of the nucleotide sequence of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene and major allergen alt a1 (Alt a 1) gene. Multiple sequence alignments of GAPDH gene (A) and Alt a 1 gene (B) of Alternaria sonchi strains CNU1001-3 and A. sonchi KC584142.1 type strain. Asterisks (*) indicate identical nucleotides and mismatched nucleotides are highlighted.