Beuve, M, Stevens, M, Liu, H. Y, Wintermantel, W. M, Hauser, S and Lemaire, O 2008. Biological and molecular characterization of an American sugar beet-infecting
Beet western yellows virus isolate.
Plant Dis 92: 51-60.


Brault, V, Périgon, S, Reinbold, C, Erdinger, M, Scheidecker, D, Herrbach, E et al. 2005. The polerovirus minor capsid protein determines vector specificity and intestinal tropism in the aphid.
J. Virol 79: 9685-9693.



Brown, J. K 2000. Molecular markers for the identification and global tracking of whitefly vector-Begomovirus complexes.
Virus Res 71: 233-260.


Brunt, A. A, Crabtree, K, Dallwitz, M. J, Gibbs, A. J, Watson, L and Zurcher, E. J 1996. Plant viruses online:descriptions and lists from the VIDE database. URL
http://bio-mirror.im.ac.cn/mirrors/pvo/vide/refs.htm [16 November 2018]
Choi, G. S, Kim, J. H, Lee, D. H, Kim, J. S and Ryu, K. H 2005. Occurrence and distribution of viruses infecting pepper in Korea.
Plant Pathol. J 21: 258-261.

Domier, L. L 2012. Family - Luteoviridae. In: Virus taxonomy:Ninth report of the international committee on taxonomy of viruses, eds. by A. M King, M. J Adams, E. B Carstens and E. J Lefkowitz, pp. 1045-1053. Elsevier, London, UK.
Dreher, T. W and Miller, W. A 2006. Translational control in positive strand RNA plant viruses.
Virology 344: 185-197.



Duffus, J. E 1960. Radish yellows, a disease of Radish, Sugar Beet, and other crops. Phytopathology 50: 389-394.
Duffus, J. E 1961. Economic significance of Beet western yellows (Radish yellows) on Sugar Beet. Phytopathology 51: 605-607.
Kim, J. H, Choi, G. S and Choi, J. K 2002. Characterization of Cucumber mosaic virus Subgroup II Isolated from Paprika (
Capsicum annuum var.
grossum) in Korea.
Plant Pathol. J 18: 6-11.

Kim, J. H, Choi, G. S and Choi, J. K 2004. Characterization of Tomato spotted wilt virus from paprika in Korea.
Plant Pathol. J 20: 297-301.

Knierim, D, Tsai, W. S, Deng, T. C, Green, S. K and Kenyon, L 2013. Full-length genome sequences of four polerovirus isolates infecting cucurbits in Taiwan determined from total RNA extracted from field samples.
Plant Pathol 62: 633-641.

Knierim, D, Tsai, W. S, Maiss, E and Kenyon, L 2014. Molecular diversity of poleroviruses infecting cucurbit crops in four countries reveals the presence of members of six distinct species.
Arch Virol 159: 1459-1465.


Kwon, S. J, Choi, G. S, Yoon, J. Y, Seo, J. K and Choi, H. S 2016. Identification of
Leonurus sibiricus as a weed reservoir for three pepper-infecting viruses.
Plant Pathol. J 32: 65-69.



Kwon, S. J, Cho, I. S, Yoon, J. Y and Chung, B. N 2018. Incidence and Occurrence pattern of viruses on peppers growing in fields in Korea.
Res. Plant Dis 24: 66-74. In Korean

Marco, S 1984. Beet western yellows virus in Israel.
Plant Dis 68: 162-163.

Mayo, M. A and Ziegler-Graff, V 1996. Molecular biology of luteoviruses.
Adv. Virus Res 46: 413-460.


Mun, H. Y, Park, M. R, Lee, H. B and Kim, K. H 2008. Outbreak of cucumber mosaic virus and tomato spotted wilt virus on bell pepper grown in Jeonnam province in Korea.
Plant Pathol. J 24: 93-96.

Nam, M, Kim, J. S, Lim, S, Park, C. Y, Kim, J. G, Choi, H. S et al. 2014. Development of the large-scale oligonucleotide chip for the diagnosis of plant viruses and its practical use.
Plant Pathol. J 30: 51-57.



Pálak, J 1979. Occurrence of beet western yellows virus in sugar beet in Czechoslovakia.
Biol. Plant 21: 275-279.

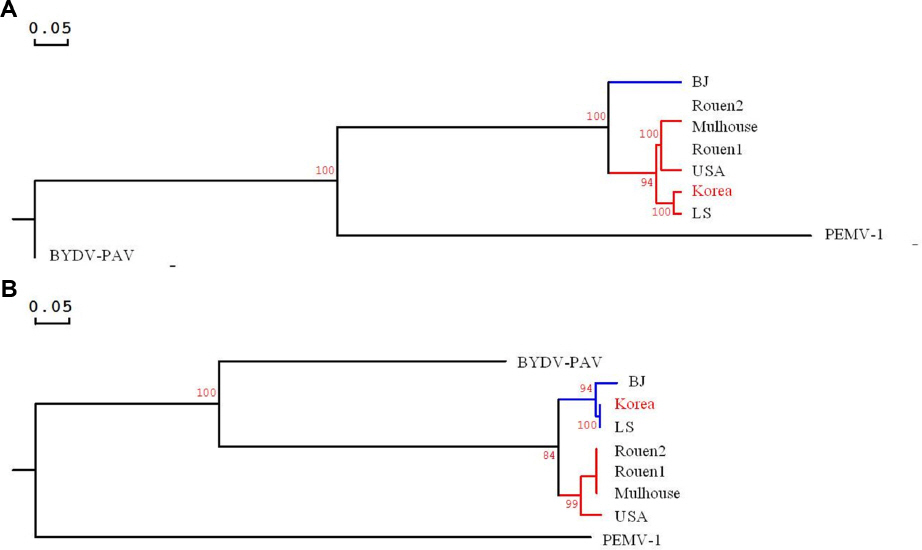
Park, C. Y, Shin, Y. G, Kim, J. S, Nam, M, Lee, J. H, Jun, E. S et al. 2011. First report of Beet western yellows virus on Capsicum annuum var. angulosum at Jinju in Korea. Res. Plant Dis 17: 463. (Abstract)
Reinbold, C, Gildow, F. E, Herrbach, E, Ziegler-Graff, V, Gonçalves, M. C, Van Den Heuvel, J F et al. 2001. Studies on the role of the minor capsid protein in transport of
Beet western yellows virus through Myzus persicae.
J. Gen. Virol 82: 1995-2007.


Russell, G. E and Duffus, J. E 1970. An aphid-transmitted yellowing virus disease of lettuce in England. Plant Pathol 19: 148-149.
Smith, H. G and Hinckes, J. A 1985. Studies on beet western yellows virus in oilseed rape (
Brassica napus ssp.
oleifera) and sugar beet (
Beta vulgaris).
Ann. Appl. Biol 107: 473-484.

Tamaki, G and Fox, L 1982. Weed species hosting viruliferous green peach aphids, vector of beet western yellows virus.
Environ. Entomol 11: 115-117.

Wang, R, Wang, N, Ye, T, Chen, S. Y, Fan, Z. F and Zhou, T 2013. Occurrence of Beet western yellows virus in Japanese hop (Humulus scandens) in China. J. Plant Pathol 95: 450.
Yuan, W, Yong, R. J, Zuo, L. Y, Du, K. T and Zhou, T 2015. First report of Beet western yellow virus on pepper in China. J. Plant Pathol 97: 400.
Zhou, C. J, Xiang, H. Y, Zhuo, T, Li, D. W, Yu, J. L and Han, C. G 2011. A novel strain of
Beet western yellows virus infecting sugar beet with two distinct genotypes differing in the 5'-terminal half of genome.
Virus Genes 42: 141-149.






 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print