The Gac/Rsm Signaling Pathway of a Biocontrol Bacterium, Pseudomonas chlororaphis O6
Article information
Abstract
Pseudomonas chlororaphis O6, isolated from the roots of dryland, field-grown commercial wheat in the USA, enhances plant health and therefore it is used in agriculture as a biofertilizer and biocontrol agent. The metabolites produced by this pseudomonad stimulate plant growth through direct antagonism of pathogens and by inducing systemic resistance in the plant. Studies upon P. chlororaphis O6 identify the pathways through which defined bacterial metabolites generate protection against pathogenic microbes, insects, and nematodes. P. chlororaphis O6 also triggers plant resistance to drought and salinity stresses. The beneficial determinants are produced from bacterial cells as they form biofilms during root colonization. Molecular control these processes in P. chlororaphis O6 involves the global regulatory Gac/Rsm signaling cascade with cross-talk between other global regulatory pathways. The Gac/Rsm regulon allows for coordinate phasing of expression of the genes that encode these beneficial traits among a community of cells. This review provides insights on the Gac/Rsm regulon in expression of beneficial traits of the P. chlororaphis O6 which can contribute to help yield enhancement and quality in agricultural production.
Introduction
The rhizosphere is a complex, dynamic, and competitive field of influence that is important for plant health, where microbial communities are nurtured by the metabolites released by plant roots. The plant growth promotion and protection properties endowed upon the plant by the rhizosphere microbes has evoked the terminologies, plant growth-promoting rhizobacteria (PGPR) and biological control agents (BCA) (Kloepper et al., 1980). Often, PGPR isolates display both growth promoting and biological control abilities. Indeed, early studies indicated that plant growth promotion was due to the suppression of deleterious microorganisms in the soil (Kloepper et al., 1980; Schippers et al., 1987). The finding of suppressive soils fits this concept, where combat among soil organisms results in greater plant growth (Weller, 1988). Such observations have fueled an interest in study of the mechanisms used by PGPR and BCA. With the current surge in studies of microbiomes, including those in the human body, we can now name PGPR/BCA as plant probiotics. In humans, probiotics are microbes that promote health through various mechanisms including direct and indirect methods to control pathogens. Thus, BCA fit into this designation. Similar to human probiotics, PGPR show diversity in metabolic catabolism that produces antibiotics and other secondary metabolites with activities including the stimulation of plant immunity, and regulation of plant development. Several BCA are commercialized (Fravel, 2005) including isolates of the spore-forming bacteria genera Bacillus spp. and the fungal genera Trichoderma spp. (Bashan et al., 2014; Pal and Gardener, 2006; Wackett, 2003; Woo et al., 2014). However, new approaches are needed to understand the way to achieve reproducible and effective results in field applications of BCA (Timmusk et al., 2015).
Biological control was initially considered to be linked to the microbial products that directly antagonize pathogen growth. These products are comparable to antibiotics, which directly suppress pathogens. However, biological control also involves a second mechanism, in which plants are protected through induction of systemic resistance against diseases (Heil and Bostock, 2002; Kuć, 1982; Pieterse et al., 2014; Yoo and Sang, 2017; Zdor and Anderson, 1992). Studies have examined the mechanisms involved in plant resistance against pathogens, and both localized and systemic activities were found (Anderson-Prouty and Albersheim, 1975; Kuć, 1982). Molecular studies on plant-microbe or plant-insect interaction studies reveal that different plant resistance pathways are deciphered. For instance, a salicylic acid dependent pathway triggered by avirulent plant pathogen. However, an ethylene/jasmonate dependent pathway is triggered upon wounding and/or herbivore challenge (Heil and Bostock, 2002; Pieterse et al., 1998). Plants have an innate immune system which, similar to that in animals (Ausubel, 2005; Jones and Dangl, 2006), involves detection of conserved molecular patterns (i.e., microbe-associated molecular patterns [MAMPs]) including bacterial and fungal cell wall polymers, and bacterial flagellin. Induction of plant resistance is an expanding field of research that is aimed at understanding the mechanisms and cellular signaling systems that are involved as well as practical field applications of PGPR/BCA.
A BCA, Pseudomonas chlororaphis strain O6 (PcO6), was isolated from the root surfaces of commercial dryland wheat at the end of the growing season in Logan, Utah, USA. Thus, this bacterium survives competition with other rhizosphere microbes and periods of soil dryness throughout the dryland growing season, and was able to mine the required essential metals, such as Fe, Zn, and Cu, from calcareous soils at pH 8.3. Subsequent studies revealed the probiotic-like traits of the isolate, starting with its direct antagonism against fungal growth through the secreted metabolites (Radtke et al., 1994). In vitro inhibitory capacity on fungal growth by the metabolites secreted by PcO6 has been demonstrated against major fungal plant pathogens, Botrytis cinerea, Pyricularia grisea, Colletotrichum gloeosporioides, Phytophthora capsici, Rhizoctonia solani, Fusarium graminearum, and Pythium spp. and Alternaria sp. Fig. 1 illustrates the inhibitory mycelial growth of Fusarium proliferatum in the presence of PcO6 correlating with secretion of antagonistic metabolites; while mycelia in the control plate eventually covers the whole agar surface, further fungal growth in the PcO6-inoculated plate is halted.
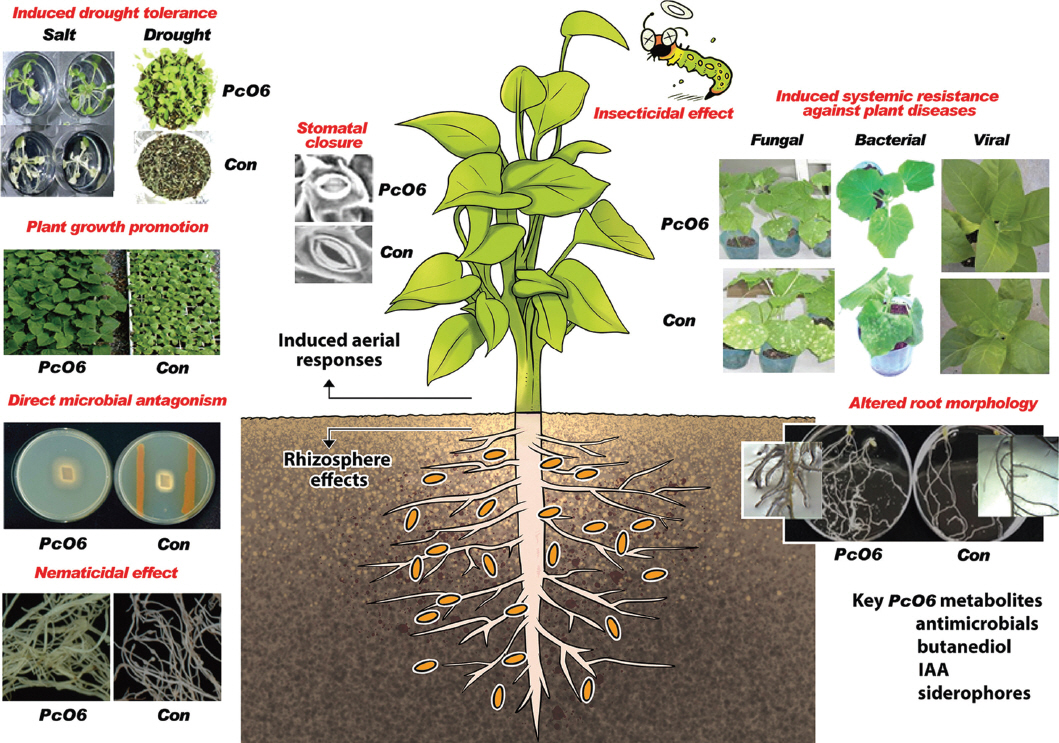
Beneficial effects of Pseudomonas chlororaphis O6 root colonization on plant health. P. chlororaphis O6 produces many metabolites that are directly antagonistic to plant pathogens, including nematodes and insects. Microbial colonization alters root morphology, increases plant growth, and induces systemic resistance against plant diseases. Control of Fe availability by P. chlororaphis O6 metabolites is an important feature in the rhizosphere. Partial stomatal closure induced by a volatile metabolite from P. chlororaphis O6, butanediol, is involved in the mechanism that induces systemic resistance against plant diseases and abiotic stresses.
PcO6 also induces systemic resistance to bacterial and fungal pathogens (Kim et al., 2004; Spencer et al., 2003), nematodes (Lee et al., 2011, 2017), and viruses (Ryu et al., 2007) as illustrated in Fig. 1. Disease symptoms are reduced for cucumber infected with Sphaerotheca fusca, Pseudomonas syringae pv. lachrymans, and Cucumber mosaic virus (Fig. 1). Other examples of pathogen resistance include systemic protection against Tobacco mosaic virus (Ryu et al., 2007) and P. syringae pv. tabaci in tobacco (Spencer et al., 2003), and against Pectobacterium carotovorum SCC1 in Arabidopsis (Han et al., 2006). Additionally, systemic resilience in Arabidopsis against abiotic stresses, such as drought and salinity, was induced by PcO6 colonization on plant roots as shown in Fig. 1 (Cho et al., 2008, 2013). Root colonization of PcO6 alleviates the intense chlorosis and wilting observed in non-colonized plants after exposure to salt. Similarly, seedlings colonized by PcO6 survived under a period of drought condition followed by rehydration whereas plants without colonization were dessicated (Fig. 1). Hosts other than plants also are affected by colonization with PcO6, because this bacterium possessed insecticidal capacity. This bacterium showed insecticidal activity on the tobacco hornworm (Rangel et al., 2016) and on nematodes (Lee et al., 2011).
Bacterial colonization of plant roots is achieved through metabolism of the array nutrients released in the exudates from plant roots. The composition of root exudate is quite similar among plants, with the major metabolites belonging to sugars, organic acids, and amino acids (Hirsch et al., 2013; Kamilova et al., 2006). The cell growth of PcO6 on chemical compounds released from plant roots has been confirmed in In vitro cultures (Martineau et al., 2014). From different experiments, PcO6 is recovered at approximately 107–8 colony forming unit (CFU)/g root, and the bacterium effectively colonizes along the length of the roots from a seed inoculum (Kim et al., 2014a, 2014c; Oh et al., 2013a).
From the previous root combination studies, the question arises as to how this bacterium orchestrates the varied metabolic processes that are required to modulate survival of other rhizosphere organisms and enhanced fitness of the host plant. As part of the answer, this review focusses on the role of a global regulatory system, the Gac/Rsm regulon, in controlling the production of the key metabolites in the protective responses to other microbes and in the protective plants (Heeb and Haas, 2001). The concerted regulation allows a community of PcO6 cells colonizing the plant root to act in unison through coordination of gene expression to control metabolite production. The Gac/Rsm network does not function alone but instead is integrated into other regulatory circuits to enhance adaption to varied stresses.
Gac/Rsm System, a Master Switch for Global Regulation
Regulation of antimicrobials
The term, global regulatory system for activation of antibiotic and cyanide (Gac) synthesis, is derived from studies of virulent and nonpathogenic pseudomonad isolates (Heeb and Haas, 2001). The proteins, GacS and GacA, interact as a classic paired sensor kinase and a response regulator system that, through phosphorelay, enables cells to respond to changing environmental conditions (Krell et al., 2010; Rodrigue et al., 2000; Stock et al., 1990). In pathogens, the Gac sensor kinase system controls virulence gene expression (Heeb and Haas, 2001). It is also found that the Gac/Rsm system of PcO6 is required for the expression of the insecticidal activity to the tobacco hornworm (Rangel et al., 2016), and to the root knot nematode (Lee et al., 2011, 2017). In addition, the Gac system is shown to be important in expression of gene expression in the inhibition of fungal growth by PcO6 (Kang et al., 2007; Oh et al., 2013a; Park et al., 2011). Presumably, a function of its direct antimicrobial control is enhanced survival in the competitive environment of the rhizosphere, where water, nutrients, and habitat must be secured in the face of other microbial invading challengers.
Table 1 summarizes the active metabolites of PcO6 that potentially are expressed in the rhizosphere and reduced in mutant lacking the Gac/Rsm regulon through a mutation in gacS. The antimicrobials of PcO6 include the volatile HCN, which is toxic to bacteria, fungi, nematodes and plants (Zdor, 2015), as well as secreted products, families of phenazines (Goodman et al., 2016; Housley et al., 2009), dialkyl resorcinols (unpublished), and the chlorinated-ring structure pyrrolnitrin (Park et al., 2011). Knockout mutations in the antimicrobial biosynthetic genes for these products reveal that these antimicrobials play either major or minor roles depending on the pathogen (Park et al., 2011).

Metabolites and structures under positive and negative regulation by the Gac/Rsm regulon in Pseudomonas chlororaphis O6
Synthesis of the highly charged metabolites, like polyamines, also plays an antagonistic role in the rhizosphere (Park et al., unpublished). In PcO6, polyamines are synthesized from arginine through pathways involving two carboxylases encoded by speA and speC. Expression of these genes is dependent on Gac/Rsm signaling, and a double speAspeC mutant is unable to inhibit the growth of several fungal pathogens. Although the fungal growth-inhibiting activity of PcO6 can be restored by the polyamines spermidine or putrescine, neither of these polyamines can restore the ability of a gacS mutant to inhibit fungal growth, indicating that these polyamines regulate the production of antagonistic chemicals in the Gac/Rsm network rather than being direct inhibitors themselves. The mechanism underlying this novel regulatory function is currently under elucidation.
Gac/Rsm regulation is activated when the cell density reaches a threshold, and by inducing this global program, the cell mass has more uniform metabolism. For the pseudomonad, the outcomes of activating this signaling include increased potential habitat and nutrients, resulting from damage to competing organisms. However, these antimicrobials may also play a role in controlling plant disease by activating ISR in the host plant; thus, these antimicrobials may act as signals between organisms from different kingdoms, and influence plant health.
Mechanics of the Gac/Rsm regulon
The Gac/Rsm regulon is far-reaching, controlling expression of the about 10% genes of the genome in P. fluorescens Pf-5, P. fluorescens SBW25, and Pc30-84 (Cheng et al., 2013; Hassan et al., 2010; Wang et al., 2013). The Gac/Rsm regulon includes expression of alternative sigma factors, each of which has its own regulatory network (Fig. 2). For instance, in PcO6, expression from rpoS is partially dependent on the Gac/Rsm system (Kang et al., 2004). If PcO6 is similar to other pseudomonads, Gac/Rsm may also regulate additional alternative sigma factors, such as the regulator of alginate biosynthesis and motility, AlgU (Fig. 2), and the iron utilization sigma factor RvdS (Frangipani et al., 2014; Hay et al., 2014). The intracellular signaling compound, cyclic-di-GMP (c-di-GMP), is also involved in this integrated network. The bacterial stress adaptation was enhanced by the global changes in gene expression that occur through regulation of these alternative sigma factors.
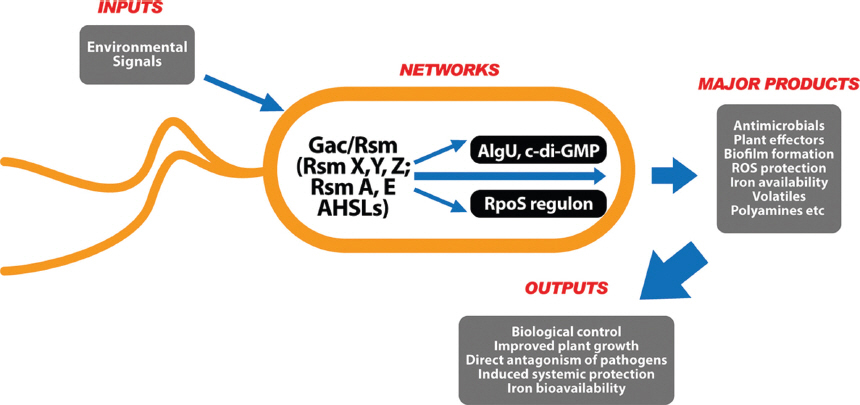
Model of the signal transduction pathways involving Gac/Rsm in Pseudomonas chlororaphis O6. Activation of the sensor kinase GacS by unknown signals triggers a phospho-relay to phosphorylate the response regulator GacA. Phosphorylated GacA positively controls the transcription of regulatory RNAs, such as RsmZ and RsmY. Overexpression of these small RNAs titrates the translational repressor proteins RsmA and RsmE, thereby making the ribosome binding site (RBS) of target mRNAs accessible. Gac/Rsm activation in P. chlororaphis O6 also leads to production of the diffusible cell signaling molecules AHSLs. The Gac/Rsm pathway networks with the regulon controlled by the alternative sigma factor RpoS. The RpoS regulon includes the transcription of genes involved in antimicrobial production (e.g., pyrrolnitrin) and protection against oxidative stress (e.g., glutathione peroxidase, Gpx), as well as modified cell envelope properties. Regulation of both Gac/Rsm and RpoS may alter the Fe sources available to the cell; GacS represses synthesis of the pyoverdine-like siderophore, whereas RpoS represses synthesis of a heme outer membrane surface receptor and heme oxidase. The Gac/Rsm regulon is also interconnected with the AlgU/c-di-GMP networks that regulate biofilm formation.
Our current knowledge of the regulatory systems governing the biocontrol potential of PcO6 is incomplete. The studies by Haas’s group on P. fluorescens and P. aeruginosa (Heeb and Haas, 2001; Humair et al., 2010; Sonnleitner and Haas, 2011), Pierson’s group on Pc30-84 (Wang et al., 2012), Girard’s group on Pc1391 (Girard et al., 2006b), and de Kievit’s group on PcPA23 (Selin et al., 2012), coupled with bioinformatic analysis of PcO6 are featured in this compilation of mechanistic pathways. Genes that may be involved in the Gac/Rsm pathway in PcO6 are shown in Supplementary Table 1.
The environmental stimulus that initiates phosphorelay from GacS in the cytoplasmic membrane to the transcription regulator GacA is unknown. However, during infection with the enteric plant pathogenic bacteria, such as Erwinia, Pectobacterium, and Dickeya spp. that causes soft rot, the plantassociated phenolics o- and p-coumaric acids and t-cinnamic acid activate the Gac/Rsm regulon (Charkowski, 2009). Yamazaki et al. (2012) obtained similar findings for activation by phenolics in P. aeruginosa.
Phosphorylated GacA induces gene expression changes in the gac regulon, in part by increasing the expression of genes encoding small regulatory RNAs, such as RsmX, RsmY, and RsmZ, which compete for binding with a family of translational repressors called the Rsm proteins (Brencic and Lory, 2009; Brencic et al., 2009; Hassan et al., 2010; Lapouge et al., 2007). The Gac regulon is kept in check by other cell signaling components. The LysR-type transcriptional regulator, PtrA, has been implicated as a master regulator of the Gac network in Pc isolate 23, as it regulates the levels of the transcriptional repressor RsmE and the small RNA RsmY (Shah et al., 2016). Two genes that condition GacS-GacA phosphorelay, ladS and retS (Sonnleitner and Haas, 2011), are also present in PcO6. There is a second potential modifying system involving RpeB and RpeA that was identified in studies on phenazine biosynthesis in Pc30-84 (Wang et al., 2012; Whistler and Pierson, 2003). The pip gene, which encodes a phenazine-inducing protein (Chancey et al., 1999; Girard et al., 2006a), that regulates PhzR, the immediate regulator of the phz biosynthetic operon, is also present in PcO6. There is also a locus for a homolog encoding the transcriptional regulator PsrA, which controls rpoS expression through the Gac/Rsm system (Girard et al., 2006b). Girard and Rigali (2011) proposed that a stressed cell uses Pip as a regulator to redirect energy through the pathways under RpoS control, rather than using carbon and nitrogen sources to generate phenazines.
Additional genes with less clear roles in Gac/Rsm regulation are also found in the PcO6 genome (Supplementary Table 1). Quorum sensing in P. aeruginosa (Coggan and Wolfgang, 2012; Sonnleitner and Haas, 2011) occurs through two mechanisms, one using acyl homoserine lactones (AHSLs) and the second using an alkyl quinolone, 2-heptyl-4 quinolone, as the signaling compounds. Although Vrf regulates both cell signals in P. aeruginosa, mutation of this gene in Pc449 does not affect the production of AHSLs (Veselova et al., 2009), and no genes with homology to the P. aeruginosa quinolone synthetic genes are detected in the PcO6 genome. However, a homolog of phrS, which encodes a small regulatory RNA that stimulates P. aeruginosa quinolone signaling, is present (Sonnleitner and Haas, 2011). Thus, the functions of Vfr and PhrS in other Pseudomonas spp. remain to be elucidated. PcO6 also possesses the rgsA gene, which encodes another small regulatory RNA that is under indirect Gac control (i.e., it is not associated with the regulatory RNAs RsmX, RsmY, and RsmZ) in P. aeruginosa and is correlated with responses to oxidative stress (Sonnleitner and Haas, 2011).
Work from Haas’s group has shown differential regulation of the genes encoding the small regulatory RNAs RsmX, RsmY, and RsmZ based on nutrition and culture phase (Humair et al., 2010). Control of expression from rsmZ may involve factors in addition to GacS-P regulation, because binding sites for integration host factor (IHF) and PsrA are also present. Another study showed regulation of these RNAs by Krebs cycle intermediates and pyruvate, suggesting linkages between the Gac regulon and basic catabolic pathways (Takeuchi et al., 2009).
Role of the quorum sensing in the PcO6 Gac/Rsm regulon
An accessory but vital arm of the gac system is the production of the intracellular and intercellular signaling compounds, acyl homoserine lactones (AHSLs). These AHSL signaling compounds diffuse from producing cells, so that all the cells in the community are in phase for coordinate expression of genes within the gac regulon. The cells then act like a population; hence, the term quorum sensing (Papenfort and Bassler, 2016). Diffusion also allows cheater cells, such as naturally occurring gacS or gacA mutants, to regain regulation by exogenous AHSLs. Detection of microbial AHSL utilization is the basis for common AHSL microbial biosensors, e.g., Chromobacterium violaceum (McClean et al., 1997).
In PcO6, two sets of genes have been shown to be associated with AHSL production. Both include the regulatory genes phzR and csaR, which condition the expression of the AHSL synthases encoded by the cognate paired inducer genes phzI and csaI (Fig. 2, Supplementary Table 1). Activation of the synthase genes enables AHLS levels to reach a threshold, and then a complex of AHLS with its regulators activates target gene expression (Fig. 2). In phenazine-inducing medium, the dominant AHSL produced by PcO6 are 3-OH C6 (hexanolyl) and 3-OH C8 (octanoyl) derivatives, although 3-OH C7 and 3-OH C10 as well as the straight chain C4, C6 and C10 AHSLs are also detected (Goodman et al., 2016). Khan et al. (2007) reported that 3-OH, C6 HSL is the most effective AHSL for phenazine synthesis in Pc30-84. The roles of these AHSLs in PcO6 metabolism have not yet been clarified.
AHSL production and corresponding phenazine synthesis in PcO6 require functional GacS/GacA phospho-relay. Production of AHSL and phenazines are control by diverse environmental factors. Nutritional quality is one such factor, and AHSL production is optimal in rich medium containing organic nitrogen sources and is low in a minimal medium limiting phenazine production (Housley et al., 2009). It is possible that these nutritional affects are related to transcriptional control of phzR through RpeA/RpeB interactions (Whistler and Pierson, 2003). Surfactants, as well as Fe and Zn levels, also regulate PcO6. The hydrophobic commercial surfactant Pluronic 25R2 was shown to increase accumulation of long chain AHSLs, whereas the hydrophilic surfactant Pluronic P123 was shown to decrease production in minimal medium (Housley et al., 2009). An experiment testing the addition of metal ions to phenazine-inducing medium showed that Fe increased phenazine and AHSL levels, while Zn decreased their levels (Goodman et al., 2016). The significance of such regulation in the control of plant stresses in the field remains to be determined. Clearly, the availability of nutrients and essential metals would by patchy and surfactant levels would be variable under field conditions, even at the microscale along a plant root. Such variations would affect AHSL levels and in turn the transcriptional control of phzR and expression from the phz biosynthetic operon. Interkingdom interactions may then come into play due to the influence of phenazines on plant health. The effects of nutrient availability on plant health could be compounded by their impact on AHSLs, because these are among the array of microbial products that activate plant defense responses. This topic has been recently reviewed (Schenk and Schikora, 2015; Schikora et al., 2016).
The synthesis of additional antimicrobials produced by PcO6, including dialkyl resorcinols, pyrrolnitrin, and HCN, are also regulated by the Gac/Rsm (Table 1). However, the intracellular signaling involved in their regulation is less clear. For instance, phenazine production is absent in a PcO6 gacS mutant but is increased in an rpoS mutant, a finding that is in agreement with observation of continued AHSL production in the absence of RpoS (Hegsted, 2010). In contrast, pyrrolnitrin production is absent in both rpoS and gacS mutants (Park et al., 2011). Monitoring HCN levels revealed a strong reduction in a gacS mutant but lesser inhibition in an rpoS mutant (Oh et al., 2013a).
In PcO6, medium composition leads to regulate the ratio of pyrrolnitrin and phenazine. For example, pyrrolnitrin production is inhibited by glucose amended during PcO6 cultivation. Consequently, production of different antimicrobials upon PcO6 growth in different nutrient was directly correlated with its extent of fungal growth inhibition (Park et al., 2011). In P. fluorescens CHA0, the Gac/Rsm pathway is linked to central metabolism, and fumarate, succinate, and pyruvate are positively triggered expression of the secondary antimicrobial metabolites (Takeuchi et al., 2009). These findings suggest the nutrient available in the nature are important in expression of antimicrobial production in PcO6, and reveals the difficulty in predicting the functions of PcO6 in the rhizosphere.
Oxidative stress and the Gac/Rsm regulon
The ability of PcO6 to survive in the oxidative stress was attempted by mutations in gacS or rpoS, because GacS regulates RpoS (Kang et al., 2004; Kim et al., 2014a). The reduced tolerance of a gacS mutant to hydrogen peroxide compared to the wild-type strain correlates with the absence of certain isozymes of catalase and peroxidase that involved in the removal of hydrogen peroxide (Kang et al., 2004). GacS mutant cells of PcO6 are under higher oxidative stress than wild-type cells. Proteomic analysis of a gacS-defective mutant revealed that transcriptional expression of katG, which encodes a major catalase/peroxidase, was down-regulated, and transcription of gpx, which encodes a glutathione peroxidase, was reduced (Kim et al., 2014b). However, the higher levels of Mn-superoxide dismutase was detected in the GacS mutant (Kang et al., 2004). These findings suggest that the Gac/Rsm regulon helps PcO6 survive under oxidative stress and other unfavorable environments, such as when challenged by antibiotics produced by other microbes (Dridi et al., 2015).
Traits Regulated Negatively by the gac Regulon
Much of the research on the Gac/Rsm regulon has been focused on the positive regulation of antimicrobials and other traits (Heeb and Haas, 2001), as is illustrated for PcO6 in Table 1. However, certain traits that are important to plant health are repressed by this regulon (Table 1). For example, production of pyoverdine-like siderophores and indole acetic acid (IAA) from tryptophan (Kang et al., 2006) are increased in a PcO6 gacS mutant. This control of siderophore production by Gac/Rsm was not restricted to PcO6, but were observed similar repressive effects of Gac on siderophore production in P. fluorescens Pf-5 (Hassan et al., 2010) and P. fluorescens SBW25 (Cheng et al., 2013).
Cell elongation and the number of flagella are also negatively regulated by GacS in PcO6. In addition, growth of the PcO6 gacS mutant is increased on certain peptides and carboxylic acids when compared to the growth of wild-type cells. Increased levels of pyoverdine-like siderophores in a gac mutant boosts cell survival under low iron conditions, such as in calcareous soils. The phenomenon also would aid pseudomonad survival in metal-contaminated conditions, because the siderophore would bind the metal and avoid its cellular toxicity (Dimkpa et al., 2012). Modified siderophore levels in rhizosphere may also impact the plant and rhizosphere microbiomes. In the rhizosphere, siderophores are important in biocontrol because their strong chelation efficacy restricts Fe availability for other microbes, including some plant pathogens (Kloepper et al., 1980). Additionally, siderophores produced by biocontrol pseudomonads can supply Fe to the plant (Nagata et al., 2013; Vansuyt et al., 2007) and induce resistance in the plant (Audenaert et al., 2002; Aznar and Dellagi, 2015; Aznar et al., 2014).
Microbial IAA production is a trait commonly associated with probiotic organisms, namely the ability to alter host development. For plants, exogenous IAA influences root development through a hometic effect, because it improves growth at low levels but inhibits elongation at higher levels. However, even if IAA production leads to stunted root growth, IAA-induced proliferation of elongated root hairs enhances the plant’s water supply and nutrient uptake (Adams et al., 2017). Although gacS mutation increases IAA production in PcO6, the effect of the Gac system on production of IAA is strain dependent. A root colonizer of Arabidopsis thaliana, P. brassicacearum produces natural GacS defective variants in the rhizosphere, and these variants have lower IAA production (Lalaouna et al., 2012). Interestingly, the PcO6 gacS mutant was auxotrophic for tryptophan, because transcription of trpE, encoding a key tryptophan biosynthetic enzyme, was significantly reduced in the gacS mutant (Kim et al., 2014a). Tryptophan is the key precursor for biosynthesis of IAA (Spaepen and Vanderleyden, 2011) and pyrrolnitrin (Salcher and Lingens, 1980), and tryptophan is derived from chorismate, which is known as the precursor of phenazines (Blankenfeldt and Parsons, 2014). Consequently, Gac/Rsm-regulated metabolism is linked to the biosynthetic pathways of several intermediates that are important for the biocontrol potential of PcO6.
Experiments using PM array plates (Park et al., unpublished) have shown that Gac/Rsm controls the catabolism of many amino acids and their di- and tri-peptides in PcO6. Increased growth on certain peptides is observed with the gacS mutants, when compared to the growth of wild-type cells. Growth in the presence of citrate and succinate was stimulated in the gacS mutant (Park et al., in press). The metabolic changes observed in the gacS mutant may be related to altered plasma membrane function and changes in cell surface charge due to modification of extracellular polymers (Anderson et al., 2005). The enhanced potential of the gacS mutant to mine for certain nutrients in the rhizosphere may be related to the ability of this mutant to colonize plant roots at wild-type levels.
Root colonization and biofilm formation
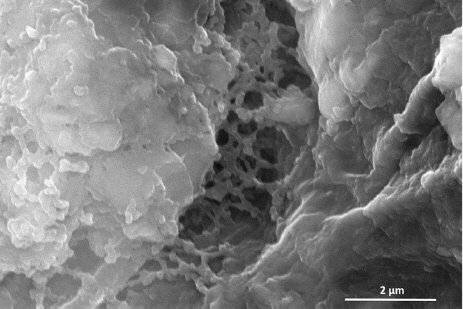
Early work with BCA demonstrated that root colonization is essential for efficacy against plant disease (Lugtenberg et al., 1991). PcO6 mutants defective in amino acid biosynthesis were poor colonizers and failed to induce systemic resistance (Han et al., 2006; Kim et al., 2014a). Induction of systemic resistance against abiotic and biotic stresses by PcO6 requires a minimum of 3 days for bacterial colonization and the production of plant-active metabolites (Cho et al., 2008). Perhaps 3 days post-PcO6 inoculation may be provided enough time to produce a strong biofilm and/or accumulate the metabolites that interact with the plant cells. PcO6 grows on plant root surfaces with the cells embedded in an extensive, layered mucilage, as shown in Fig. 3. Gac/Rsm regulation was reported to act central role on biofilm formation in pathogenic P. aeruginosa isolates (Frangipani et al., 2014) and in Pc isolate 30-84 (Wang et al., 2015). To understand biofilm formation in PcO6, we have studied two of the processes involved, cell motility and extracellular matrix production.
Motility
Motility is involved in the initial stages of biofilm formation (Davey and O’Toole, 2000). When grown on low-strength agar plates to induce swimming or swarming, the diameters of the gacS mutant colonies are largest, and colony diameter is reduced by complementation of this mutation (Fig. 4). Increased swimming mobility in PcO6 is gacS-dependent but rpoS-independent (Kim et al., 2014b). Enhanced swimming in PcO6 gacS mutant correlates with more polar flagella in the gacS mutant. Increased transcript abundance of fliQ, which encodes a protein involved in flagellin export, flhF, which encodes a protein involved in flagella placement, and fleQ, which encodes a transcriptional regulator, are observed in the gacS mutant (Kim et al., 2014b).
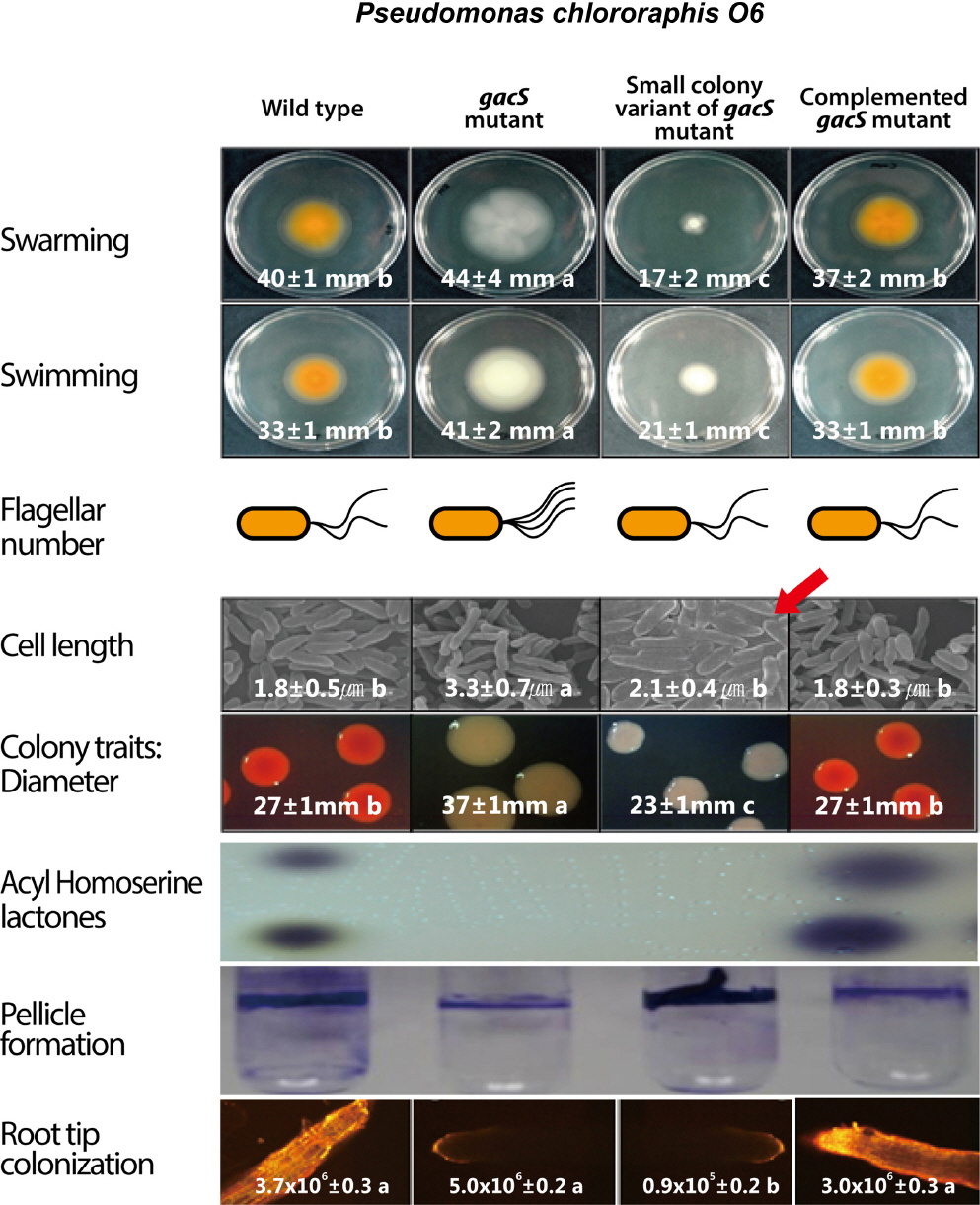
Comparative traits of wild-type Pseudomonas chlororaphis O6, a gacS mutant, and a naturally occurring small colony variant (SCV). Mutation of gacS increases the swimming and swarming motilities, correlated with increased numbers of flagella. Cells of the gacS mutant strain are also longer. SCV cells are highly hydrophobic, accounting for their increased aggregation, leading to thicker biofilm formation (shown here as a pellicle) and lesser motility than the gacS cells. The biofilms of both gacS and SCV cells form in the absence of AHSLs and phenazines. Confocal imaging of bean roots shows that wild-type cells and complemented gacS mutant cells, but not gacS mutant or SCV cells, produced fluorescence on the root surface. However, all cell types colonized the root surface.
In P. aeruginosa, FleQ is an adaptable regulator, functioning as an activator of flagella formation and either as a repressor or activator of biofilm matrix polymers, dependent on cellular status (Baraquet and Harwood, 2016; Hickman and Harwood, 2008). The switch for biofilm formation is governed by the level of the intracellular signaling compound c-di-GMP (Baraquet and Harwood, 2016), and the network of genes expressed at high c-di-GMP levels establishes the biofilm lifestyle. Earlier work on P. fluorescens F113 (Martínez-Granero et al., 2012) proposed that FleQ was controlled by c-di-GMP and the alternative sigma factor AlgU, which is regulated by the translational repressors RsmA and RsmE in the Gac/Rsm network. Thus, cooperative interactions exist between the cdi-GMP and Gac/Rsm networks (Fig. 2). Modifications to the cell surface polymers in PcO6 gacS mutant, perhaps through FleQ activation, are evident from direct measurement of cell stickiness by atomic force microscopy and ethylene glycol droplet contact angle measurements. The cells from a gacS biofilm are more hydrophilic than those from a wild-type biofilm (Anderson et al., 2005). The increased length of gacS cells compared to wild-type cells (Anderson et al., 2005; Kim et al., 2014b) may also be integral to its biofilm-forming potential.
The effect of the gacS mutation on the promotion of swarming in PcO6 contrasts with observations in other bacteria. In P. fluorescens Pf-5, motility was reduced in the gacS mutant (Hassan et al., 2010). The reduced swarming of Pf-5 gac mutants correlates with a loss of expression from ofa, which encodes enzymes that produce a novel surfactant (Hassan et al., 2010). Consequently, we believe that wild-type PcO6 fails to produce surfactant under growth conditions tested. Enhanced swarming of PcO6 occurs when surfactants from another microbe, such as Pseudomonas lurida (unpublished), or commercial products, such as Pluronics, are provided (Housley et al., 2009). Thus, PcO6 can be an effective root colonizer, taking advantage of metabolites produced by other organisms to boost its own survival in the rhizosphere. Studies with P. aeruginosa isolates suggest that swarming, like swimming, involves hyper flagellation as well as retractable type IV pili (Kohler et al., 2000). The formation of type IV pili in PcO6 may be affected by a gacS mutation due to decreased expression from cpaC, the gene encoding a protein required for type IV pilin transport (Kim et al., 2014a). Analogous to P. aeruginosa, in which expression of type IV pilins represses swarming (Kuchma et al., 2012), such changes in the PcO6 gacS mutant may contribute to enhanced swarming (Fig. 4).
Biofilm formation
Staining of PcO6 biofilms with crystal violet to measure biomass showed plasticity in biofilm formation, related to nutrient availability and culture age (Kim et al., 2014c). For instance, substitution of sucrose for mannitol in minimal medium supports a thicker biofilm of gacS mutant cells (Kim et al., 2014c).
Proteomic studies of the gacS mutant revealed changes in the functions of the cellular envelope compared to wild-type cells that are likely associated with the observed plasticity in biofilm formation (Kim et al., 2014c). Most notable are the reduced levels of the major outer membrane protein OprF, the CpaC transport channel that is involved in type IV pilus formation, and the peptidoglycan-binding protein LysM. P. aeruginosa oprF mutant shows enhanced biofilm formation through increased extracellular polysaccharides, which are related to elevated levels of RsmZ and the intracellular signaling compound c-di-GMP (Bouffartigues et al., 2015). The cell envelope changes caused by the gacS mutation in PcO6 may also overlap with the RpoS regulon (Fig. 2). The cell envelope changes in the PcO6 rpoS mutant include reduced levels of the outer membrane secretory pore TolC and the inner membrane polyamine translocator PotA (Oh et al., 2013b). Of additional interest is that the rpoS mutant overexpresses two proteins, heme oxidase and the outer membrane heme receptor CirA (Oh et al., 2013b). Thus, mutations in both gacS and rpoS in PcO6 may maximize Fe acquisition. However, Fe acquisition in GacS and RpoS may be different, a GacS-regulated process involving increased chelation by a pyoverdinelike siderophore and a second mechanism under RpoS control that may tap into alternative heme sources.
The interactions between the Gac/Rsm and RpoS regulons also suggest that polyamine levels are modified in gacS mutant cells. We predict that import of polyamines from external sources is restricted by RpoS-dependent changes in PotA, and internal synthesis from arginine is limited by GacS-dependent regulation. Recent review indicated that cellular polyamines are linked oxidative stress resistance, biofilm formation, and iron acquisition in bacterial pathogens (Shah and Swiatlo, 2008). Similarly, the potential regulatory role of polyamines in PcO6 (Park et al., unpublished) extend the array of genes regulated in this biocontrol pseudomonad.
Small colony variant of PcO6 gacS mutant
The gacS mutant spontaneously generates small colony variants (SCV), which grow on solid medium as raised colonies with small diameters (Kim et al., 2014c), that are correlated with a dramatic increase in cell surface hydrophobicity (Anderson et al., 2005). Biofilms of wild-type cells produce contact angles with droplets of ethylene glycol of 18±6°, and biofilms of gacS cells produce similar contact angles (28±7°), whereas SCV biofilms produce contact angles of 114±12°. SCV cells aggregate in liquid culture and form biofilms, including pellicles (Fig. 4), that are denser than those of the gacS mutant (Anderson et al., 2005). The cell elongation trait observed in the gacS mutants was also observed in the SCV (Anderson et al., 2005). In contrast, expression of flhF and fliQ, and flagella number in the SCV was similar to those of wild-type (Fig. 4). However, the colony size in swimming and swarming assays on agar medium remains small (Fig. 4), which we presume is due to intense aggregative interactions between cells. The SCV cells initially attach and colonize plant roots, remaining recoverable as the seedlings mature, although to a lesser degree than the gacS mutant (Fig. 4).
SCV formation is also observed in P. aeruginosa strains (Coggan and Wolfgang, 2012; Drenkard and Ausubel, 2002) and Pc30-84 (Wang et al., 2015). These variants show modifications in the genes that increase the levels of c-di-GMP, consistent with a battery of phenotype changes focused on a loss of motility and but a gain of strong biofilm formation (Römling et al., 2013; Valentini and Filloux, 2016). The changes that occur in cells defective in Gac/Rsm signaling demonstrate a connection with the c-di-GMP signaling network (Fig. 2) and the plasticity in biofilm formation control. Mann and Wozniak (2012) previously remarked on the flexibility of biofilm formation in P. aeruginosa and the role of extracellular matrix modification in this process. SCV of P. aeruginosa show enhanced resistance to antibiotics, adding to the persistence of this bacterial pathogen (Malone, 2015). A similar role for SCV formation in the rhizosphere seems feasible.
Induced systemic tolerance by PcO6
The mechanism in planta that leads to induced systemic resistance due to root colonization by PcO6 is not yet well understood. However, speCific antimicrobials that have been isolated and purified from PcO6, i.e., phenazines, pyrrolnitrin, and pyoverdine- like siderophores have been shown to be effectors of induced systemic responses in the plant. The earliest work with PcO6 colonization of tobacco revealed that resistance to the wild fire bacterial pathogen involved systemic signaling in the plant through the ethylene/jasmonate signaling pathway (Spencer et al., 2003). Such a connection to the ethylene/jasmonate pathway upon pathogen protection has also been observed in root colonization with other beneficial microbes (Kupferschmied et al., 2013). A novel metabolite with a structure related to salicylate has been identified as one of the effectors of induced resistance in tobacco (Park et al., 2008). The plant stress cell signaling compound jasmonate is associated with induction of a gene encoding the osmolyte galactinol, which accumulates during PcO6-mediated alleviation of drought stress (Cho et al., 2011). PcO6-induced resistance against plant pathogens that invade through the stomates could involve stomatal closure by butanediol produced by PcO6 (Cho et al., 2012). Stomatal closure is also induced by signaling through ABA, salicylic acid, nitric oxide, and hydrogen peroxide (Cho et al., 2012). Recent research by other groups has suggested that AHSLs induce resistance through the synergistic interaction of pathways that include salicylic acid and oxylipids (Schenk and Schikora, 2015; Schikora et al., 2016). These findings demonstrate that the response to a bacterium that causes no physical damage overlaps with the systemic defense pathways associated with pathogen attack, including the classical salicylic acid and ethylene-jasmonate signaling pathways (Heil and Bostock, 2002; Pieterse et al., 1998).
Transcriptome studies with plants colonized by PcO6 show the stealthy nature of their association, as there are no major changes in host gene expression following colonization (Cho et al., 2011). The bacterium exists on the root surface without triggering any major changes in resistance gene expression. Instead, the colonization process involves changes to plant metabolism that promote more rapid and effective resistance gene expression upon challenge. This process is termed priming (Conrath et al., 2015) and occurs in response to both biotic and abiotic challenges. The array of transcripts that accumulate after priming and challenge include many involved in the response to pathogen attack, as well as to those encoding proteins and osmolytes associated with protection against water stress (Cho et al., 2010, 2011). Such activation confirms the plant-level observations of reduced disease symptoms upon pathogen challenge and better health after exposure salt and water stress (Fig. 1).
Recent studies with Bacillus isolates (Timmusk et al., 2015) point to the establishment of thick bacterial biofilms on the root surface as significant factors in drought stress protection. Thus, the mucilage covering the root surface generated by PcO6 (Fig. 3) likely aids the plant under drought conditions because the root surface cells now have zones protected by a hydrated gel. The gel could also increase the levels of bioactive PcO6 metabolites by limiting their diffusion into the rhizosphere. This effect may work in conjunction with the responses in aerial tissues, involving stomatal closure, triggered by the volatile butanediol produced by PcO6 biofilms on the root surface (Cho et al., 2008). Studies using the gacS mutant of PcO6 are in progress to determine whether colonization of the root by bacterial cells that do not produce butanediol and the other metabolites (Table 1) under Gac/Rsm control can similarly protect the plants.
Conclusion
The network of the Gac/Rsm system in PcO6 conveys the abilities needed to adapt to different challenges and promote survival while supporting plant health by priming stress responses. When present at a crucial cell density, the microbe activates the Gac/Rsm network to change its nutrient use and produce protectants rather than add cell mass. The Gac/Rsm system bridges with other mechanisms controlling gene expression, speCifically those involving the cell signaling molecule c-di-GMP and the RpoS regulon. Activation of the Gac/Rsm network in PcO6 generates antimicrobials that may help maintain plant root colonization and protect the plant from microbial pathogens. The secretion of antimicrobials and volatiles by PcO6 influences microbiomes and plant health in the rhizosphere. At the plant root surface, the bacterium produces a protective biofilm over the root surface that functions as a physical shield and a water-withholding gel. Here, many of the Gac/Rsmregulated metabolites initiate systemic resistance responses in the plant. However, although the Gac/Rsm system is a major positive regulator of metabolism that is beneficial to the host plant, inactivation of the Gac/Rsm system in PcO6 does not eliminate root colonization. However, inactivation triggers the generation of metabolites and changes in cell surface structures that also promote plant welfare. Thus, the Gac/Rsm network coordinates the metabolism of the microbial population to promote both survival of the bacterium and the performance of the host plant.
Supplementary Table 1
Acknowledgement
The research was supported by a grant from the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01250602)” of the Rural Development Administration, Republic of Korea and by a recent grant from NIFA-USDA to AJA (2011-2015).
References
Notes
Conflicts of Interest
The authors declare that they have no competing and commercial interests in this work.