First Report of Fusarium Wilt Caused by Fusarium proliferatum on Safflower
Article information
Abstract
Wilt disease appeared the first in greenhouse-grown safflower (Carthamus tinctorius) in Jeonju, Korea. With the advancement of the disease, the infected plants were withered and died. In order to investigate the causal organism of this symptom disease, fungus was isolated from the infected plants and cultured on potato dextrose agar medium. The fungus showed the white or orange colony color with aerial mycelium. Macroconidia were from falcate to straight, usually 3–5 septate with 38.0–66.7× 2.9–4.4 µm. The fungus was inoculated to a new safflower plant and caused the same wilt. With morphological characters and pathogenicity results, sequence analyses (internal transcribed spacer ribosomal DNA and translation elongation factor 1α) suggested that, the isolated fungus is Fusarium proliferatum. This is the first report of Fusarium wilt disease caused by F. proliferatum on safflower in Korea.
Introduction
Safflower (Carthamus tinctorius) is cultivated herbaceous annual plant of the family Asteraceae. Safflower has been widely cultivated in Korea, India, China, Egypt, Southern Europe, North America, and Australia (Lee, 1980). Safflower seeds contain α-linoleic acid, which used as cooking oil and clinically for the treatment of cataclasis, osteoporosis and rheumatoid arthritis in Korea (Kang et al., 1999; Kim, 1992; Lee et al., 2002). In addition, its flower has been utilized for folk medicine as an analgesic, antithrombotic and antihypertensive crude drug, as well as natural dye and food colorants (Han, 1988).
It has been reported that main pathogens on safflower are Alternaria alternata (leaf spot), Botrytis cinerea (gray mold), Colletotrichum acutatum (Anthracnose), Fusarium oxysporum (Fusarium wilt), Phytophthora cactorum (Phytophthora root rot), Puccinia carthami (rust), Sclerotium rolfsii (collar rot), and Sphaerotheca fuliginea (powdery mildew) worldwide (Farr and Rossman, 2016). Recently, we observed wilt symptoms on safflower grown in green house in Jeonju, Korea, thus leading to identify the causal agent of the wilt disease in this study.
Isolation of the pathogen
Several wilted safflower were observed in May, 2015 in Jeonju, Korea. The symptoms appeared as brown discoloration of leaves on the lower part of stem (Fig. 1A). The infected plants were wilted with sudden dropping of leaves (Fig. 1B). Severely infected plants were covered by whitish fungal mass (arrow) (Fig. 1C). Small pieces of stem from the diseased plant were sterilized with 75% ethanol and 1% sodium hypochlorite for 30 seconds followed by 2 times washing with sterile distilled water. They were placed onto water agar and incubated at 25°C. After 2 days, hyphal tips of the emerging fungus were transferred onto potato dextrose agar (PDA; Difco, Sparks, MD, USA) medium to pure culture, and incubated at 25°C. The Isolated pathogen was kept in 20% (v/v) glycerol at –70°C for further study.
Morphological characteristic
Fungal isolates were grown on PDA at 25°C for 7 days. Morphological characteristics of the isolate were observed under a light microscopy (DM5500B; Leica, Wetzlar, Germany). The isolated fungi H226 produced whitish colony with aerial mycelia on PDA, becoming tinged in orange color (Fig. 2A). The mycelial growth of isolated fungi H226 on PDA for 7 days were founded to be optimum at 23°C–25°C. Microconidia were produced in chains or in false heads (Fig. 2B). Macroconidia were subfalcate to needlelike, usually 3–5 septate, hyaline, smooth, 38.0–66.7×2.9–4.4 µm (Fig. 2C). Microconidia were clavate with a truncate base, 0–1 septate, hyaline, smooth, 5.3–14.7×2.9–3.5 µm (Fig. 2D). Conidiophores were cylindrical, often proliferated, branched or unbranched, with mono- and poly-phialides. No clamydospores were observed. On the basis of morphological features, the isolated fungus was assumed as F. proliferatum (Table 1) (Ichikawa and Takayuki, 2000; Nelson et al., 1983).
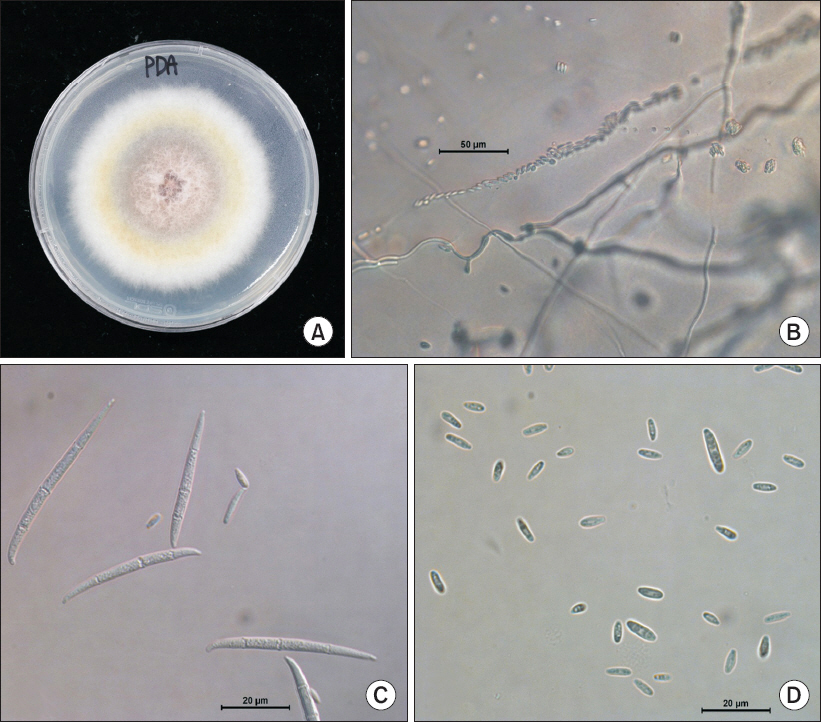
Morphology of Fusarium proliferatum isolated from safflower. (A) Fungal colony on a potato dextrose agar (PDA) after 7 days of incubation at 25°C. (B) Chain of microconidia. (C) Macroconidia. (D) Microconidia of isolated pathogen.
Pathogenicity assay
Pathogenicity assay was conducted on 3 weeks old safflower plants in green house. The conidial suspension of H226 was prepared from 5-day-old culture grown on potato dextrose broth and adjusted to 4×106 conidia/ml. The conidia suspension (10 ml) was inoculated via drenching onto healthy safflowers grown in plastic pots, and then the inoculated plants were placed under green house. Inoculated plants exhibited same symptom such as discoloration and wilting of leaves 1 week after inoculation, but no symptom were shown on the control plant. The fungus was successfully re-isolated from the artificially inoculated plants, fulfilling Koch's postulates.
Sequence analysis
For further identification, sequences obtained from ribosomal DNA-internal transcribed spacer (rDNA-ITS) region and translation elongation factor 1α (TEF) gene region of the isolated fungus were analyzed. Genomic DNA was extracted from mycelia using cell lysis solution (Qiagen, Maryland, MD, USA), RNase A solution (Biosesang, Seongnam, Korea), protein precipitation solution (Qiagen), and Isopropanol (Fisher Scientific Korea Ltd., Seoul, Korea). The region of rDNA-ITS and TEF gene was amplified from genomic DNA using primer pairs ITS1/ITS4 and EF-1/EF-2, respectively (Geiser et al., 2004; Hsuan et al., 2011; White et al., 1990). Each amplified fragment was sequenced and compared with all fungal sequences available in the GenBank database (National Centre for Biotechnology Information; http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Fusarium ID (http://isolate. fusariumdb.org/blast.php) using BLAST program. With our sequences, reference sequences of Fusarium spp. and related fungi were used for the phylogenetic analysis. A neighbor-joining tree was constructed by the Tajima-Nei parameter model of MEGA 6 (Saitou and Nei, 1987; Tamura et al., 2013).
PCR products (523 bp) of isolate H226 by primer ITS1 and ITS4 were sequenced and deposited GenBank (accession No. KU218561). This sequence data was compared with other fungal sequences available in the GenBank database using BLAST search tool. The BLAST search showed that the sequence derived from H226 isolate was 99% similar to the ITS sequence of F. proliferatum (accession Nos. KP407870, GU980584, KJ634671). In addition, PCR products (678 bp) amplified by primer EF-1 and EF-2 were sequenced and deposited in GenBank (accession No. KU218560). This sequence data showed 99% of similarity to the TEF sequence of F. proliferatum (accession Nos. FJ603514, KP732060, JX174033). BLAST search using FUSARIUM-ID also showed 99% of similarity to TEF sequence of F. proliferatum (accession Nos. FD_01380, FD_01379, FD_01776). Phylogenetic analysis of the ITS and TEF region sequences revealed that H226 isolate belongs to group of F. proliferatum (Fig. 3, 4). Taken together, our results suggested that H226 isolate is F. proliferatum, based on morphological characteristics, sequence analysis and pathogenicity to the host.
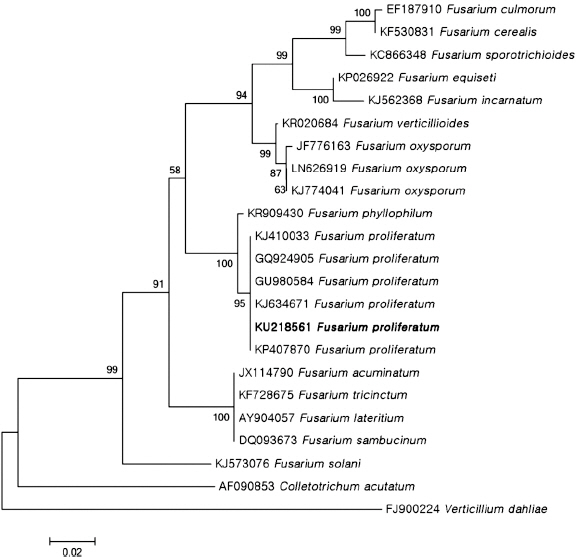
Phylogenetic analysis of sequence of the internal transcribed spacer ribosomal DNA (rDNA) region of the Fusarium proliferatum H226 with closely related strains retrieved from GenBank. The tree was constructed based on the neighbor-joining method with 1,000 replicates. The numbers above the branches represent the bootstrap value. The fungus identified in this study is boldfaced.
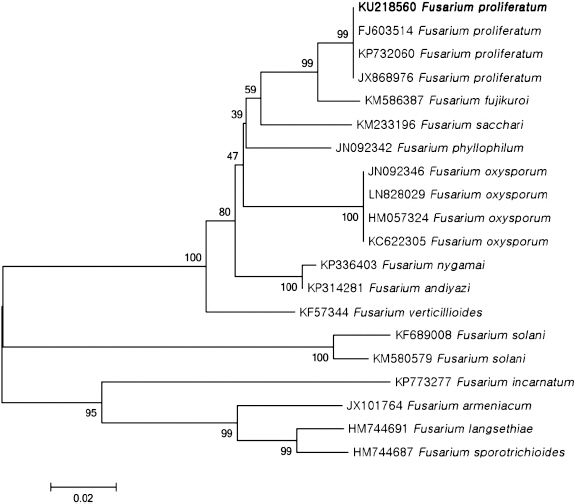
Phylogenetic analysis of sequence of the translation elongation factor 1α region of the Fusarium proliferatum H226 with closely related strains retrieved from GenBank. The tree was constructed based on the neighbor-joining method with 1,000 replicates. The numbers above the branches represent the bootstrap value. The fungus identified in this study is boldfaced.
To date, there have been no reports of the occurrence of the wilt disease caused by F. proliferatum on safflower in Korea. According to the Systematic Mycology and Microbiology Laboratory Fungus-Host Database, one species of Fusarium, F. oxysporum, was reported on safflower (Farr and Rossman, 2016). To our knowledge, this is the first report of F. proliferatum on safflower in Korea.
Acknowledgement
This study was supported by a grant (project No. PJ0101180) funded by National Institute of Agricultural Sciences, Rural Development Administration, and by a 2016 Postdoctoral Fellowship Program of the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.

