Plant diseases are health problems in plants primarily caused by various pathogens such as fungi, bacteria, viruses, and nematodes. While it is difficult to pinpoint the exact time when humans began to recognize that plants can get sick, the awareness of plant diseases has existed in human history for a long time. The scientific understanding that plant diseases are caused by biological pathogens began to develop in 1863 when Anton de Bary proved that potato blight is caused by a biological pathogen (
Matta, 2010). Traditionally, plant diseases were diagnosed visually, but this method heavily relied on experience and had limitations in differentiating complex and diverse pathogens. Serological tests are also widely used for diagnosing plant diseases, but they are primarily limited to viruses. With the advancement of molecular biology in the 20th century, methods for diagnosing by analyzing DNA/RNA were developed (
Martin et al., 2000). This method is very accurate and can distinguish different pathogens that show the same symptoms, but it is time-consuming, costly, and requires specialized skills and equipment. Recently, to overcome these limitations, biochemical diagnostic methods using volatile compounds or the introduction of spectroscopic techniques have been adopted (
Farber et al., 2019;
Riu et al., 2022).
Meanwhile, with the advancement of computer vision technology, various machine learning/deep learning algorithms have been developed to analyze plant disease images and diagnose them quickly (
Jackulin and Murugavalli, 2022). These methods, trained with large volumes of plant disease photo data, provide consistent and accurate diagnoses and can process vast amounts of data quickly, making them useful for detecting plant diseases in large-scale farms. However, they require high-quality training data and can suffer in diagnostic performance if the data is insufficient or biased. Moreover, diagnosing based solely on images has limitations in detecting characteristics of host plants or complex environmental factors that may not be fully apparent in the image data. Nevertheless, the introduction of the latest artificial intelligence (AI) technology, especially large multimodal models (LMMs), has helped overcome these limitations in the fields of life sciences and dentistry (
Huang et al., 2023;
Wang et al., 2023). In plant disease diagnosis, models like GPT-4 can analyze text and image data simultaneously, enabling deeper contextual understanding and comprehensive diagnostics (
Qing et al., 2023). Through this, AI can go beyond simple image classification to comprehensively analyze various data, such as environmental factors, climate change, and the physiological state of plants, thereby enhancing the accuracy and reliability of plant disease diagnosis.
This study has developed an AI plant doctor system that recognizes plants and lesions and diagnoses plant diseases using GPT-4-vision, an LMM capable of text and image recognition (
Fig. 1). The system was developed using OpenAI's application programming interface (API), runs on the Streamlit package, and is programmed in Python 3. One method to utilize GPT as an AI plant doctor involves employing prompt engineering to limit the tasks the GPT model can perform, thereby recognizing GPT as an expert in plant diseases. These prompts, coded as instructions, are used to create user-defined GPT models and endow GPT with unique characteristics. To prevent the extraction of prompts, they are set to be private. However, it is disclosed that the prompts are designed to discourage excessive use of chemical pesticides and emphasize sustainable practices. Furthermore, in the case of queries unrelated to plant diseases, users are prompted to ask questions related to plant diseases.
Fig. 1.
The concept of Artificial Intelligence Plant Doctor incorporated with a knowledge base. The GPT4-vision-based AI plant doctor receives input as an image and text. The AI is incorporated with a knowledge base, including lists of plant diseases and pesticides. For the output, the GPT offers specific diagnoses, causal explanations, intervention methods, prevention tips, and further resources for the provided information.
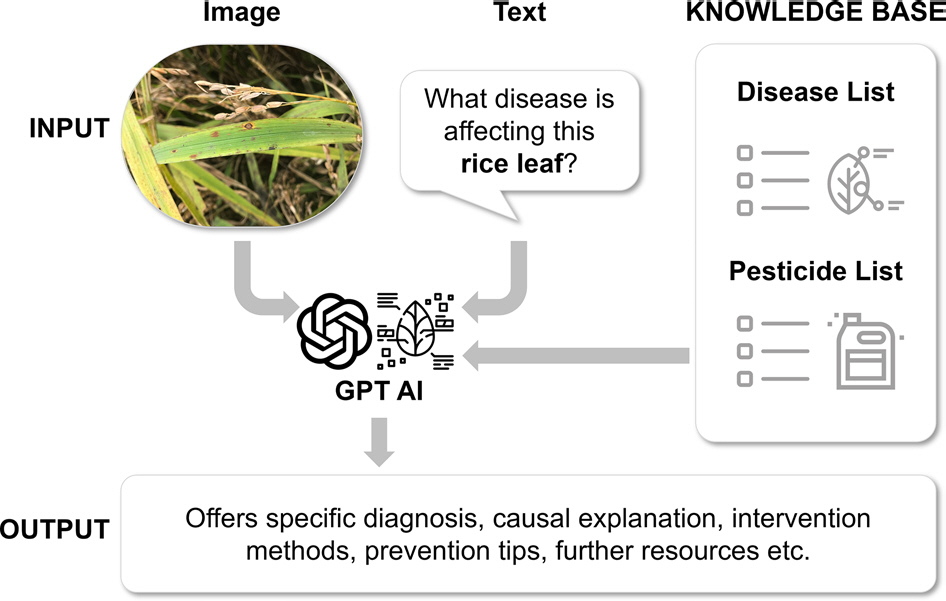
To differentiate from the standard GPT-4 model, it utilizes a vast knowledge base including 1,420 host plant taxa and 2,462 pathogen taxa listed in the 6.1 edition of the Korean Plant Disease Index (
Kim et al., 2023) and 37,467 cases of pesticides available for each plant recorded in the Rural Development Administration's Pesticide Safety Information System. This data is stored in the knowledge base in the form of comma-separated values (CSV) files in a table format. The plant disease list includes the Korean, English, and scientific names of the host plants, along with the Korean and English disease names and scientific names of the pathogens. The pesticide list includes the plant name, disease name, pesticide name, generic name, trade name, mammalian toxicity, fish toxicity, mode of action, dilution rate, application rate, optimal application timing, application method, safety application timing, safe application frequency, formulation, and company name. In conjunction with prompts, the knowledge base serves as a method to endow the user-defined GPT with unique characteristics, enabling GPT to respond to user queries within a limited scope of knowledge. When a user inputs a host plant, limiting the pathogens that can infect the host through a list of plant diseases acts as a safeguard to enhance GPT's accuracy and reduce the probability of incorrect answers.
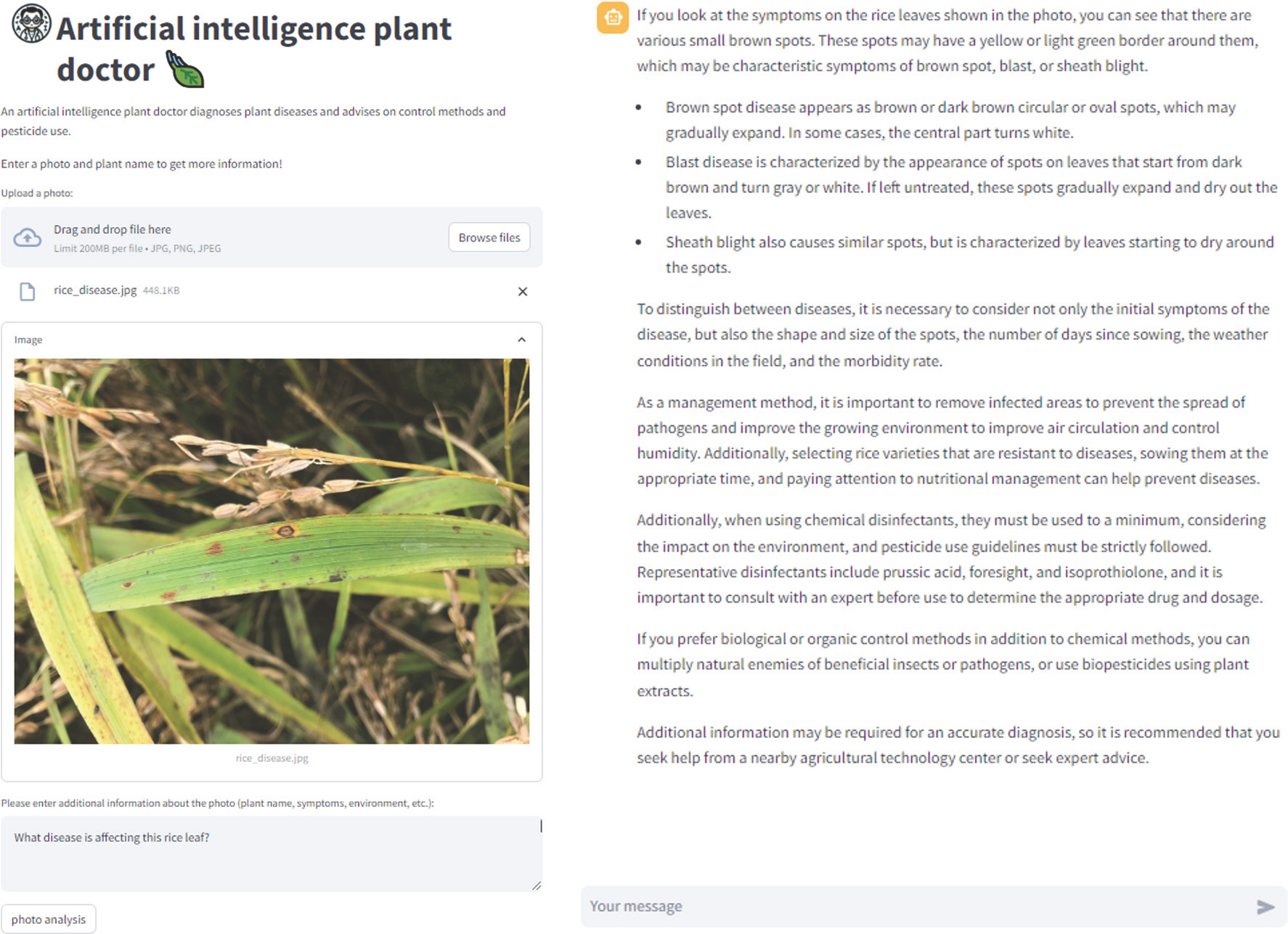
To enhance the accuracy of diagnosis, the system allows text input of the host plant's name, description of symptoms, and environmental factors. For instance, in the case of rice blast, which recently became a major problem in South Korea, it typically shows acute, watery dark brown to chronic fusiform symptoms, and occurs frequently in low temperatures and high humidity during summer (
Song et al., 2022). Thus, if a user provides text about the shape or color of lesions, and the crop's growing environment, along with the host plant, the system analyzes both image and text data to comprehensively assess the type of pathogen. Based on this information, the system can analyze images and related text data to recognize complex patterns and accurately identify various types of plant diseases (
Fig. 2). As a result, the AI plant doctor informs the user about the description of the lesions and the pathogens that could potentially infect the plant based on the photo queried by the user in a conversational manner, and provides advice on pesticide treatment in response to further inquiries from the user. Additionally, for user convenience, it provides links to plant disease-related websites such as the National Crop Pest Management System, Pesticide Safety Information System, and Korean Plant Disease Index.
Fig. 2.
The interface of AI Plant Doctor website translated into English (from Korean). The interface is designed to assist users in diagnosing and managing plant health issues by combining user-submitted photos and text information with artifical intelligence analysis.
Despite the significant potential of the AI plant doctor system developed in this study, there are several important issues to consider. Firstly, unlike traditional machine learning/deep learning models, the pre-trained GPT model lacks standardized methods for measuring accuracy and reliability. This poses considerable challenges in objectively evaluating and benchmarking the model's performance. To mitigate this drawback, this research attempted to enhance the model's accuracy and reliability by adding a knowledge base. However, this approach also necessitates a rich and detailed text database describing lesions and the environmental conditions of plant disease occurrence. Collecting and managing such data is a time-consuming and resource-intensive task, but it will improve the system's performance. Furthermore, there are questions about the system's ability to distinguish between plant diseases and non-biological stresses, such as nutrient deficiency, cold damage, or heat stress. Since various stress responses in plants can be similar, accurately differentiating them is a complex issue. This requires a high degree of accuracy and situational awareness, and with the current level of technology, such distinctions may not be perfect. These issues represent challenges faced by this research and will be a primary focus in future research and development efforts.
In conclusion, the fusion of AI technology and plant disease diagnosis represents a significant advancement in modern agriculture and plant pathology research. This innovation enables farmers and researchers to diagnose plant diseases quickly, providing insights necessary to respond to the spread of plant diseases. The progress in AI is impacting not only plant disease research but also sustainable agricultural practices, crop improvement, and the enhancement of global food security. Researchers can now use large-scale data for more precise diagnostics and understand the complex patterns and correlations necessary to comprehend the emergence and spread of plant diseases. This opens a new era in the field of agriculture, holding the potential to offer a better future for humanity and the environment through continuous technological innovation and research.








