Leatherleaf fern (
Rumohra adiantiformis) is a beautiful ornamental plant used in cut flower arrangements due to its extended shelf life and symmetrical pyramidal-shaped fronds (
Kloepper et al., 2013). Its function brings high economic value as an important world export commodity (
Nair et al., 2020) and Indonesia is one exporter country of the ornamental leaf. However, one of the fern cultivation companies in Magelang Central Java, Indonesia (GPS coordinate: -7.3726, 110.2190) currently reported a leaf blight outbreak which prominently threatened their leaf production and hugely increased loss up to 50% during rainy season. Previous outbreak reported in the leatherleaf fern cultivation there was anthracnose caused by
Colletotrichum sp. (
Sumardiyono et al., 2011) which occurred more than 10 years ago. It is said that the anthracnose disease has been well managed currently which did not cause harmful effect to the plants and production. However, “new disease outbreak” was reported in the area, different from anthracnose disease which greatly influenced the leaf production and export.
The emerging disease is a leaf blight with initial symptom was reddish brown color spots appeared from edge of leaf parts, gradually turned to blackish-brown necrotic lesions which spread to entire leaf, leaf blight and withered. Ultimately, the leaves could not be either harvested or sold. Before the “new disease outbreak” was reported, there was no significant report on it in Indonesia. In 2011, some pathogens causing necrotic in leatherleaf fern were reported, which were caused by
Cercospora sp.,
Stigmina sp., and
Alternaria sp. (
Damaiyani and Lestari, 2011). Due to the significant problem which needs swift diagnose, this study conducted Koch-Postulate and molecular identification using some marker genes to identify the pathogen. It is found that the leaf blight outbreak was caused by genus of
Neopestalotiopsis.
In Indonesia,
Neopestalotiopsis has not been reported as plant pathogen yet, however in many countries this pathogen was reported as a causal agent for many important diseases. An outbreak of strawberry leaf spot and fruit rot caused by
Neopestalotiopsis spp. was reported in Florida (
Baggio et al., 2021; Baggio and Peres, 2020).
Neopestalotiopsis siciliana sp. nov. and
N. rosae were reported to cause stem lesion and dieback on avocado in Italy (
Fiorenza et al., 2022),
N. rosae is caused to leaf spot and crown rot of strawberry in Turkey (
Erdurmuş et al., 2023),
N. guajavicola sp. nov. causing a new leaf spot on
Psidium guajava in Pakistan (
Haq et al., 2023), and first report of leaf brown blight caused by
N. formicarum was reported in Taiwan (
Lin et al., 2022). Moreover in Indonesia,
N. zimbabwana was isolated from Basidiomycete fungi
Xylaria sp. (
Hermawan et al., 2021), but was not in the plant. Based on our best knowledge, this is the first report of Neopestalotiopsis leaf blight, as an emerging concern on leatherleaf fern in Indonesia. Due to leatherleaf fern is an essential Indonesian export commodity and global trading product, this report is not only important for researcher and academic purpose, but also for practitioners and quarantine officers.
In this study, infected leaves with typical leaf blight symptom were collected from a fern plantation in Magelang, Central Java, Indonesia. Leaf samples were isolated using potato dextrose agar (PDA; Merck, Darmstadt, Germany) medium and incubated at room temperature (approximately 28°C) for 3 days. Fungal grew in the medium were re-cultured and maintained as a pure culture. Morphological identification was conducted to observe the colony shape, color and conidial shape. A pathogenicity test using mycelial plugs was conducted on the surface of fern leaves that had been sterilized with 70% EtOH. Four leaves used as replications were arranged in the 15 cm petri dish with moist paper filter at the base. Some parts of leaves were pricked with a needle and inoculated by mycelial plug of the fungal culture. After 24 hr the mycelial plug was removed and incubated at room temperature every day until the symptom appeared.
DNA extraction for molecular identification was carried out using fungal mycelia grown on PDA media. Extraction followed the Mini Kit (Plant) Genomic DNA Protocol (Geneaid, New Taipei, Taiwan) under the kit protocol and amplified using three pairs of multigene primer set of ITS region, ß-tubulin and translation elongation factor (tef1-α) gene markers, which were: and ITS5 (GGA AGT AAA AGT CGT AAC AAG G) ITS4 (TCC TCC GCT TAT TGA TAT GC) (
Singh et al., 2021); T1 (AAC ATG CGT GAG ATT GTA AGT) and Bt2b (ACC CTC AGT GTA GTG ACC CTT GGC) (
Glass and Donaldson, 1995); EF1-728F (CAT CGA GAA GTT CGA GAA GG) and EF2 (GGA GGT ACC AGT GAT CAT GTT) (
O'donnell et al., 1998). Temperature adjustment of the polymerase chain reaction (PCR) cycle according to the MyTaq HS Red Mix protocol with an annealing temperature of 52°C for ITS5-ITS4, 52°C for T1-Bt2b and 52°C for EF1-728F-EF2. PCR reaction was performed in 50 μl mixture of 25 μl of 2× DNA
Taq polymerase (MyTaq HS Red Mix; Bioline, London, UK), 19 μl miliQ sterile water, 2 μl of each marker forward and reverse, and 2 μl of DNA template in T100 Thermal Cycler machine (Biorad, CA, USA). The PCR product was visualized with the PowerPac Universal BIORAD electrophoresis machine using 1% agarose gel with DNA florosafe staining at 100 V for 30 min. The DNA bands on the agarose gel were then observed using a High-Performance UV Transilluminator. The amplicon then sent to 1st BASE Pte Ltd Singapore for sequencing. Due to the sequencing results of the three multigene primer sets, the sequences were concatenated and the phylogenetic tree analysis was carried out using the MEGA X program (
Kumar et al., 2018). Similarities were checked for nucleotide sequence homology were searched using BLAST (Basic Local Alignment Search Tool) software from NCBI (National Center for Biotechnology Information;
www.ncbi.nlm.nih.gov).
The emerging disease reported in the leatherleaf fern plantation mainly has reddish to brown lesions from the leaf edge spreading to entire leaf causing leaf dried. The symptom was obviously seen in the field (
Fig. 1), and finally they could not be either used or sold. Based on the isolation result, one dominant culture was gained (namely PK.1 isolate) which has cottony mycelia, white color in top surface (
Fig. 2A) and white to slightly peach color in the bottom surface (
Fig. 2B). Microscopically, the fungal isolate has conidia that typically seen as pestalotiopsis-like conidia (
Fig. 2C) which were 20.1-25.0 µm in length and 5.2-7.3 µm in width, hyaline with pigmented median cells, rough-walled, fusoid form, straight to slightly curved, five cells (four septa) with three median cells in light brown color, and the septa were darker. Pathogenicity test resulted the isolate PK.1 caused leaf blight (
Fig. 3) while control leaves treated by sterile water did not show any symptom. The confirmation was done by re-isolation and same culture were gained in this process. These results approved that PK.1 isolate was the pathogen causing leaf blight in the leatherleaf fern.
Fig. 1.
Neopestalotiopsis leaf blight symptom on leatherleaf fern in field.

Fig. 2.
Culture of Neopestalotiopsis sp. PK.1 isolated from leatherleaf fern on potato dextrose agar medium. (A) Upper colony surface, (B) lower colony surface, and (C) fungal conidia.
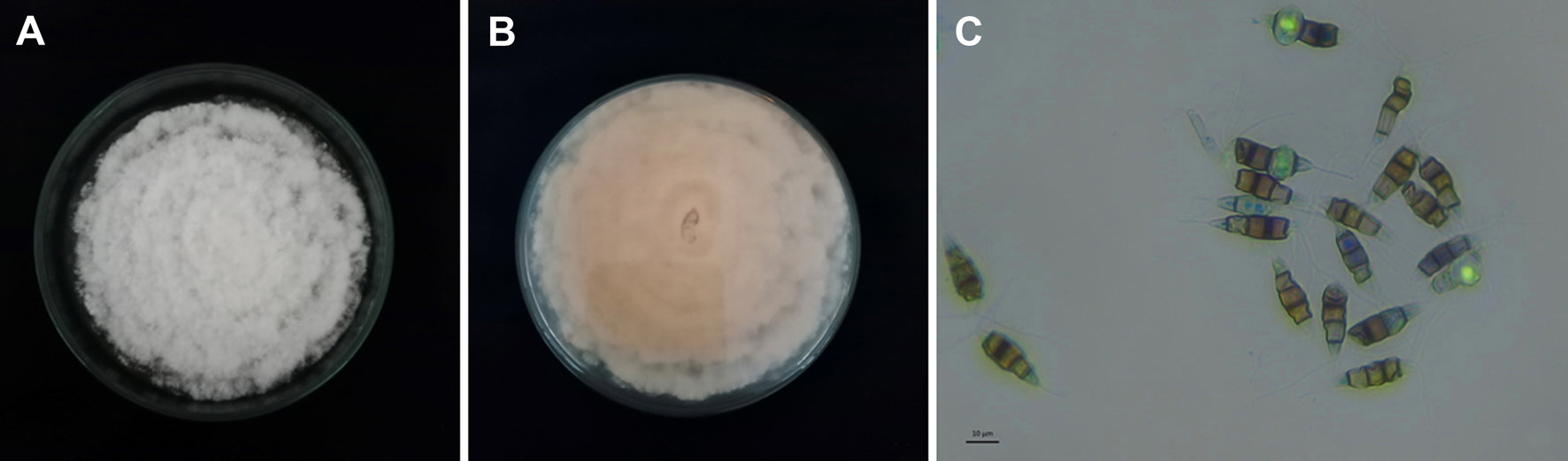
Fig. 3.
Symptom after pathogenicity test. (A) Two days after inoculation, (B) 7 days after inoculation, and (C) control 7 days after inoculation with sterile water.

Molecular identification was carried out by multigene analysis using three set of primers from ITS region, ß-tubulin, and tef1-α gene markers (
Gualberto et al., 2021). By using the three markers, some species of
Neopestalotiopsis, such as
N. rosae, N. javaensis, N. mesopotamica, and
N. formicarum were concatenated and aligned with PK.1 to gain accurate species name (
Table 1). However, phylogenetic tree analysis (
Fig. 4) showed that PK.1 isolate was closely related to some species of the genera, although the analysis has been done using three primer sets commonly used for
Neopestalotiopsis (
Gualberto et al., 2021). Based on the phylogenetic tree using multigene analysis, it is found that the PK.1 isolate from leatherleaf fern in Indonesia posed a different branch with other reported
Neopestalotiopsis spp. Therefore, it is concluded that PK.1 isolate, the pathogen causing leaf blight on leatherleaf fern in Indonesia was
Neopestalotiopsis sp. based on ITS region, ß-tub, and tef1-α gene markers.
Fig. 4 showed that
Neopestalotiopsis sp. PK.1 isolate laid in different branch compared to other
Neopestalotiopsis spp., but whether it was novel species or specific from Indonesia, needs further research to prove. The isolate used in this study has been deposited in GenBank and could be accessed with accession number of OR905551 (ITS), OR899817 (b-tubulin), and OR899816 (TEF).
Table 1.
Sequence used in the phylogenetic analyses with GenBank accession number
|
Taxa |
Strain number |
Host |
Country |
ITS |
TUB |
TEF |
|
Neopestalotiopsis sp. |
PK.1 |
Rumohra adiantiformis (this study) |
Indonesia |
OR905551 |
OR899817 |
OR899816 |
|
N. rosae
|
CBS 124745 |
Paeonia suffruticosa
|
USA |
KM199360 |
KM199430 |
KM199524 |
|
CRM-FRC |
Fragaria sp. |
Mexico |
MN385718 |
MN268529 |
MN268532 |
|
CBS 101057 |
Rosa sp. |
New Zaeland |
KM199359 |
KM199429 |
KM199523 |
|
N. javaensis
|
MFLUCC 12-0594 |
Vitis vinifera
|
France |
KX816905 |
KX816933 |
KX816874 |
|
CBS 257.31 |
Cocos nucifera
|
Indonesia |
KM199357 |
KM199457 |
KM199548 |
|
N. mesopotamica
|
CBS 336.86 |
Pinus brutia
|
Iraq |
KM199362 |
KM199441 |
KM199555 |
|
N. samarangense
|
MFLUCC 12-0282 |
Syzygium samarangense
|
Thailand |
JQ968609 |
JQ968610 |
JQ968611 |
|
N. piceana
|
CBS 254.32 |
Cocos nucifera
|
Indonesia |
KM199372 |
KM199452 |
KM199529 |
|
N. clavispora
|
CBS 447.73 |
Unknown |
Sri Langka |
MH860736 |
KM199443 |
KM199539 |
|
N. formicarum
|
CBS 115.83 |
Unknown |
Cuba |
KM199344 |
KM199444 |
KM199519 |
|
CBS 362.72 |
Unknown |
Ghana |
KM199358 |
KM199455 |
KM199517 |
|
N. saprophytica
|
CBS 115452 |
Litsea rotundifolia
|
China |
KM199345 |
KM199433 |
KM199433 |
|
Ps. cocos
|
CBS 272.29 |
Cocos nucifera
|
Indonesia |
KM199378 |
KM199467 |
KM199553 |
|
Ps. indica
|
CBS 459.78 |
Hibiscus rosa-sinensis
|
India |
KM199381 |
KM199470 |
KM199560 |
Fig. 4.
Phylogenetic tree of PK.1 isolates analysed from concatenated sequences of ITS (ITS4/ITS5), ß-tubulin (T1/Bt2b), and elongation factor (EF1-728F/EF2) gene markers. The tree was built using the maximum likelihood method with the Kimura model and tested using 1,000 bootsraps replications. Scale bar showed the genetic distance.

Genus
Neopestalotiopsis was initially introduced by
Maharachchikumbura et al. (2014) to accommodate
Pestalotiopsis-like taxa having versicolorous median cells and indistinct conidiophores. Up to date, 91 species of
Neopestalotiopsis are recognised (Mycobank 2023;
https://www.mycobank.org/page/Home) and still well-developed along with advance taxonomic study especially to get the references isolates. In complex species such as
Neopestalotiopsis, it is known that the ITS region alone cannot distinguish between closely related species (
Fiorenza et al., 2022; Prasannath et al., 2021) therefore multigene analysis was needed to identify the fungal species. However, this study showed that more gene markers and number of isolates are needed to study deeper of the species which currently get important status as plant pathogen.
Neopestalotiopsis pathogens are often considered as weak pathogen that infects plants with weak conditions. However in Florida,
N. rosae on strawberry plants showed much higher infection severity which the symptoms appeared in several types of tissues (
Baggio et al., 2021; Baggio and Peres, 2020). In the case of strawberry plants, Neopestalotiopsis leaf spot disease symptoms are similar to leaf scorch disease and ordinary leaf spot. While in fruit tissue,
Neopestalotiopsis disease symptom was very similar to anthracnose symptom. The main difference is the development of black spores on fruit lesions of
Neopestalotiopsis fruit disease, while the fungus that causes anthracnose has salmon-colored spores.
Neopestalotiopsis crown rot symptoms are difficult to distinguish from
Phytophthora and anthracnose crown rot infections (
Holland, 2021). Recently in Turkey,
N. rosae was also reported to cause leaf spot and crown rot of strawberry after molecular identification by using primers generated from ITS and tef1-α gene (
Erdurmuş et al., 2023). While
N. mesopotamica was first reported as the pathogen of strawbbery root and crown rot in Ecuador (
Hidrobo-Chavez et al., 2022).
N. formicarum recently has been reported to cause some plant disease problem such as on rubber tree in Thailand (
Pornsuriya et al., 2020), on
Kadsura coccinea, a traditional Chinese medicinal plant (
Zhonghou et al., 2023), and on guarana plant, a medicinal plant from The Brazilian Amazon (
Gualberto et al., 2021).
This study observed that Neopestalotiopsis leaf blight spread widely in the leatherleaf plantation field which eventually cause leaf dried and death. Three primer sets have been used to molecularly identify the species of pathogen, however based on the markers, it was shown that N. formicarum, N. rosae, and N. mesopotamica were closely related genetically, therefore deeper study on Neopestalotiopsis is compulsury to comprehend this “relatively new” species. Despite of the species name, this study showed the potential risk caused by fungal pathogen Neopestalotiopsis sp. lead an emerging concern in leatherleaf fern in Indonesia and biosecurity globally because the pathogen infected an important export commodity. This study also reminded that the current status of Neopestalotiopsis sp. as plant pathogen is significant to cause an outbreak of “a new emerging plant disease”. Further concern that Neopestalotiopsis could possibly infect other crops and has wide host range are also essentially required.












