Genetic Structure of Macrophomina phaseolina Populations, the Causal Agent of Sesame Charcoal Rot Disease in Iran
Article information
Abstract
Charcoal rot disease, caused by the fungus Macrophomina phaseolina, is one of the most important diseases of Sesame (Sesamum indicum) all over the world. However, the population biology of M. phaseolina is poorly understood. In this study, M. phaseolina isolates from five different regions of Iran (Khuzestan, Fars, Bushehr, Hormozgan, and Kohgiluyeh & Boyer-Ahmad provinces) (n=200) were analyzed for genetic variation using inter simple sequence repeats marker. In total, 152 unique haplotypes were identified among the 200 M. phaseolina isolates, and gene diversity (H=0.46–0.84) and genotypic diversity were high in each of the regions. The structure analysis clustered five Iranian populations into two distinct groups, the individuals from group 1 were assigned to the Bushehr population and the individuals from Khuzestan, Fars, Hormozgan and Kohgiluyeh & Boyer-Ahmad were aggregated and formed group 2. The results matched with genetic differentiation and gene flow among regions. Analyses of the distribution of gene diversity within and among five Iranian populations were 61% and 39%, respectively. Our results showed that infected seeds are thought to be the dominant mechanism responsible for the spreading of the pathogen in southern parts of Iran. In summary, it is essential to have local quarantine and prevent seed exchanges between geographical populations to restrict the dispersal of pathogen over long distances and provide certified seeds in Iran.
Introduction
Sesame (Sesamum indicum L.), one of the most economically important oilseed crops, belongs to the Pedaliaceae family and is mostly cultivated in tropical and subtropical regions of the world (Chattopadadhyay et al., 2019; Wang et al., 2021). Tanzania, India, Myanmar, and China are the main sesame producing countries, among which Iran being the 16th sesame producer in the world (Food and Agriculture Organization of the United Nations, 2023). Sesame plants are considered as significant oilseed crops with economic value, but these plants are mostly affected by biotic stress factors induced by viruses, bacteria, fungi, nematodes, and insect pests which can limit sesame production (Yadav et al., 2022), Among biotic stress, charcoal rot disease is major limitation affecting commercial sesame cultivation of arid and semiarid regions in the world and can cause seriously reduced sesame quality and yield (Min and Toyota, 2019; Teklu et al., 2022). Macrophomina phaseolina (Tassi.) Goid is a soiland seed-borne fungal pathogen that has a broad geographic distribution and causes damping off, seedling blight, charcoal rot, and infects any part of the host plant at any stage of growth from plant seed germination to maturity stage (Farr and Rossman, 2023; Kumar and Dubey, 2023; Meena et al., 2018; Roy et al., 2008). Yield losses have been reported of 5–100% in grain yield and protein contents in the world (Farr et al., 2024; Marquez et al., 2021; Vashisht et al., 2023; Win and Oo, 2017). This polyphagous pathogen has a wide host range, infects over 500 plant species in more than 100 families including monocot and dicot plant hosts, that considered as a major threat and distributed in most sesame-producing regions of the world, especially in dry and low rainy years (Farr and Rossman, 2023; Kumar and Dubey, 2023; Lodha and Mawar, 2020; Marquez et al., 2021; Sarr et al., 2014). Obviously, this pathogen's host diversity and geographical dispersal are responsible for its better survivability (Khamari et al., 2022; Kumar and Dubey, 2023; Lodha and Mawar, 2020; Vashisht et al., 2023).
This phytopathogenic fungus belongs to anamorphic ascomycetes, a member of the family Botryosphaeriaceae, and is characterized to produce pycnidia on infected stems that are responsible for secondary spread by releasing a huge number of pycnidiospores in some plants (Ghosh et al., 2018). In contrast, there is only an unconfirmed report of the sexual stage named Orbilia obscura of M. phaseolina (Ghosh et al., 2018). This necrotrophic pathogen can survive as microsclerotia in plant debris or soil for up to 15 years, and for up to 3 years as microsclerotia in symptomatic seeds or as mycelium in asymptomatic seeds (Ghosh et al., 2018; Londoño, 2022; Vashisht et al., 2023). It infects hosts tissues or seeds serving as a primary source of inoculum, distributes via seed and soil, permitting it to thrive under high temperatures (32–35ºC) and low soil moisture (below 60%) and ultimately reduces the absorption of water and nutrients, causes wilting and death, makes it hard to control M. phaseolina because of its durability as sclerotia in the soil and plant debris (Elmerich et al., 2022; Ghosh et al., 2018; Kumar and Dubey, 2023; Vashisht et al., 2023).
In the last two decades, the genetic structure of M. phaseolina populations has been investigated with molecular markers e.g., amplified fragment length polymorphic DNA (AFLP) (Linhai et al., 2011; Mayék-Pérez et al., 2001; Saleh et al., 2010), restriction fragment length polymorphisms in the rDNA region (Purkayastha et al., 2006), random amplified polymorphic DNA (RAPD) (Mahdizadeh et al., 2012; Tančić Živanov et al., 2019; Yousef, 2021), and inter simple sequence repeats (ISSR) (Khamari et al., 2023; Khamari et al., 2022; Mahdizadeh et al., 2012; Yousef, 2021). Obtained results were inconsistent in these studies, while some studies disclosed a significant correlation between the geographical locations, genetic diversity and host plants of the isolates (Mahdizadeh et al., 2012; Reznikov et al., 2019); others were not capable to confirm these findings. Aboshosha et al. (2007) and Su et al. (2001) discovered host-specific RAPD genotyping, whereas Das et al. (2008) detected a correlation between RAPD markers and the geographical location of different isolates. Mayék-Pérez et al. (2001) found AFLP fingerprints of M. phaseolina isolates were associated with geographical locations and different hosts. In contrast, a number of investigators have revealed no association between DNA genotypes, host origin and geographic origin in M. phaseolina populations. Omar et al. (2007) and Purkayastha et al. (2006) distinguished no correlation between RAPD marker and their geographic origin. Almeida et al. (2003) unveiled no host-specific M. phaseolina isolates using RAPD markers. Reyes‐Franco et al. (2006) could differen-tiate genetically between the isolates from their geographic origin and AFLP marker. It should be noted that clustering of data could not obviously distinguish isolates based on their pathogenicity, morphological characteristic and host origins (Marquez et al., 2021). In several studies, the distribution of M. phaseolina genotypes has been found to be independent of sampling position and host (Khan et al., 2017; Tančić Živanov et al., 2019).
Charcoal rot disease will be more problematic in agricultural areas under climate change scenarios such as higher temperatures accompanied by long periods of drought (Purkayastha et al., 2006; Saleh et al., 2010). The climate of southern Iran is characterized by hot, dry, rainless summers and short, cool winters that contribute to its prevalence and cause a major reduction in sesame production (Ershad, 2009). In parts of Iran, M. phaseolina is reported causing charcoal rot disease on a broad range of host plants such as chickpea, sunflower, common bean, soybean, sesame and leading to major yield losses (Ershad, 2009; Salahlou et al., 2016). Regardless of the many research efforts to control charcoal rot disease, the management strategies of M. phaseolina remain a challenge (Kumar and Dubey, 2023; Marquez et al., 2021). Responsible management of diseases caused by M. phaseolina is necessary, since the significance of this soil-borne fungus lies not only in the damages it causes but also in the influences it has on the environment because of unsupportable management practices (Vimal et al., 2017). Honestly, M. phaseolina has been present for decades in Iran; still little information is available about the genetic structure of the Iranian M. phaseolina populations that infect sesame. In an earlier study, a small number of isolates of M. phaseolina were isolated from three different regions in Iran and compared using RAPD and ISSR markers (Mahdizadeh et al., 2012; Salahlou et al., 2016), which were not appropriate representative for this study in Iran. Wholly, Knowledge of the genetic structure of M. phaseolina populations is valu-able in the development of disease management strategies (McDonald and Linde, 2002). In this study, we were able to obtain a large collection of M. phaseolina isolates from five important sesame-growing regions in southern Iran, assayed and compared five populations of M. phaseolina. The objective of this work was to study the genetic structure of M. phaseolina populations by determining the genetic diversity and differentiation of these populations using ISSR marker, which were recovered from major sesame-growing regions in five different geographic locations of Iran.
Materials and Methods
Field surveys and sample collection.
During the summer of 2019 and 2020, surveys were conducted across five sesame fields in the major sesame production regions of southern Iran in which wilting, foliar yellowing with black stems were recorded, comprising Khuzestan (Ramhormoz), Fars (Jahrom), Bushehr (Dashtestan), Hormozgan (Bandar Abbas), and Kohgiluyeh & Boyer-Ahmad (Ghachsaran) provinces (Fig. 1). Sampling was carried out using hierarchical strategies to investigate genetic structure as described by McDonald et al. (1995). Pathogen populations were collected from one sesame field in each province during the summer until early autumn and sampling locations were more than 100 km apart (Fig. 1). The symptomatic samples of root, crown and stem were transported to the laboratory in pre-sterilized polyethylene bags and kept in the refrigerator until diagnos-tic laboratory isolations.
Isolation and morphology identification.
Isolates of M. phaseolina were obtained from sesame plants showing typical symptoms (Fig. 2). Infected tissues were chopped into small sections, 4–6 mm, with a sterile scalpel from the necrotic lesions boundary, sterilized in alcohol (70%) for 30 sec and af-terward, 1% sodium hypochlorite (NaClO) for 45 sec, washed in sterile water, dried on sterile paper towels. The pieces were placed onto a potato dextrose agar (PDA) medium (12 g/l potato extract, 11 g/l dextrose, 1.2% agar) amended with chloramphenicol (100 μg/ml) to prevent bacterial con-tamination. The plates were incubated for 1 week at 30°C and observed daily growth (Moslemi et al., 2023; Pickel et al., 2020). It should be noted that, besides the infected tissues, the seeds of the infected plants were cultured in the same way. Purification of Macrophomina isolates was carried out by hyphal tip technique on water agar medium (Moslemi et al., 2023). Macrophomina species were identified using morphological characteristics, according to the taxonomic keys of the genus Macrophomina described previously by Crous et al. (2006) and Sarr et al. (2014). For long-term storage of isolates, many toothpicks were autoclaved twice, and placed on pure culture of the fungus Macrophomina on PDA medium for the colonization and formation of microsclerotia. After 2 weeks, the toothpicks dried at room temperature under laminar airflow and placed in a sterile 2 ml tube at 4°C (Moslemi et al., 2023).
DNA extraction and molecular identification.
For molecular identification, 50 out of 200 isolates from five different regions of Iran Iranian were randomly selected which were isolated in Moslemi et al. (2023) (Supplementary Table 1). Mycelium plugs from isolates were grown on PDA for 5 days, and then a 5 mm disc from a 5-day-old pure culture was transferred to 100 ml PD broth in a 250 ml flask and grown for 1 week on a rotary shaker at 30°C; the mycelium was harvested in a sterile 2 ml microcentrifuge tube, and lyophi-lized. Total DNA was extracted using the cetyltrimethylammonium bromide technique according to the procedure of Murray and Thompson (1980). The quality of the genomic DNA was checked using 1.2% agarose gel in 0.5× TAE buffer, visualized to stain with ethidium bromide solution and photographed under UV light (GelDoc; Bio-Rad Laboratories, Hercules, CA, USA). The genomic DNA is stored at -20°C until used. To confirm the identification of M. phaseolina isolates, the sequencing of 50 isolates was performed using pair of primers EF1-728F/EF2R (Carbone and Kohn, 1999; Jacobs et al., 2004) and ITS1/ITS4 (White et al., 1990) were used to am-plify translation elongation factor 1-α (tef1-α) and ITS-rDNA regions, respectively. Protocol for polymerase chain reaction (PCR) amplifications and DNA sequencing was previously described in detail in Moslemi et al. (2023). PCR products were purified from agarose gels using a PCR purification kit (MBI Fermentas, Berlin, Germany) and DNA sequencing was performed in both directions by Macrogen Company (Seoul, Korea).
Genetic diversity analysis.
Seven previously described ISSR primers, ISSR10, ISSR09, ISSR02, PcMs, P4, ISSR5, and ISSR3 (Khamari et al., 2022; Salahlou et al., 2016) amplified in each isolate. PCR amplifications were carried out in volumes of 20 μl comprising 1 μM primer (CinnaGen Co., Tehran, Iran), 0.4 μM dNTPs (MBI Fermentas), 1× Dream Taq buffer (MBI Fermentas), and 0.5 U Dream Taq DNA polymerase (MBI Fermentas). The PCR cycle parameters were: initial denaturation step at 96°C for 2 min; followed by 35 cycles at 96°C for 30 sec, primer annealing according to Salahlou et al. (2016), extension at 72°C for 1 min and a final extension step at 72°C for 5 min. The quality of PCR products was assessed on a 1.2% agarose gel, stained with ethidium bromide, viewed and photographed under UV light (GelDoc; Bio-Rad Laboratories).
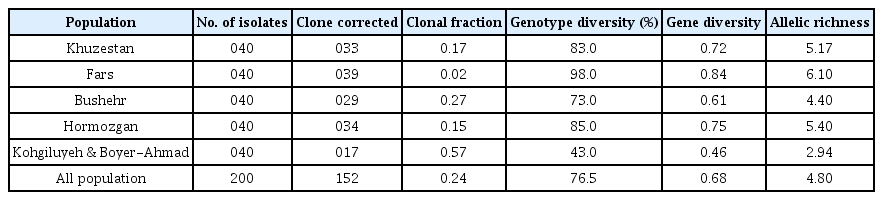
Genetic variation was calculated by measures of genotype and gene diversity in each population. A clone corrected data set in which only a single representative haplotype was used for subsequent analysis using the program Genodive 2.0 (Meirmans and Van Tienderen, 2004).
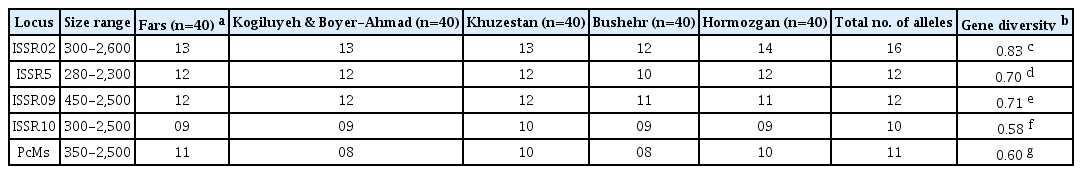
Gene diversity and the number of alleles for each population and locus, allele frequencies and the percentage of polymorphic loci (Nei, 1973) were evaluated using the program POPGENE version 1.32 (Yeh et al., 1997). The gene diversity for each locus was subjected to analysis of variance (ANOVA) using SAS v.9.1 software (SAS Institute, Inc., New York, NY, USA) and the means were compared with Duncan's multiple range test (P≤0.05). The percentage of maximum possible diversity (G/N) was measured by dividing the genotypic diversity by the number of isolates (Chen et al., 1994). Allelic richness is calculated using the program FSTAT (Goudet, 1995). Genetic differentiation among populations was estimated as FST using the program GenAlEx 6.41 (Peakall and Smouse, 2006). The effective migration rate (Nm) or gene flow was assessed based on the average FST using the formula Nm = (1 - FST) / 2 FST (Slatkin, 1993). An analysis of molecular variance (AMOVA) was carried out to calculate the hierarchical distribution of population subdivision at regional scales using the program GenAlEx 6.41 (Peakall and Smouse, 2006).
The program STRUCTURE 2.3.4 (Pritchard et al., 2000) was used to analyze population genetic structure of M. phaseolina and recognize individuals that were admixed or had migrated. The number of populations ranged from K=1 to K=10 and each isolate was assigned to one or more of K clusters. Structure Harvester version 0.6.9 was used for the graphical representation and summation of the STRUCTURE runs (Earl and VonHoldt, 2012). The implemented ΔK approach was used to recognize the optimal number of the population structure of M. phaseolina in the data set (Evanno et al., 2005).
Result
Morphological and molecular identification.
In total, 200 M. phaseolina isolates were obtained from seeds (10 isolates) and tissues (190 isolates) of sesame plants (Fig. 2) in southern Iran based on morphological characteristics described previously by Crous et al. (2006) and Sarr et al. (2014). The M. phaseolina species were efficiently isolated in late September and sampled from naturally infected sesame fields located in five major provinces (Khuzestan, Fars, Bushehr, Hormozgan, and Kohgiluyeh & Boyer-Ahmad) in the south of Iran. The Iranian populations came from five major provinces, separated by mountains, seas and deserts, which are characterized by sesame cultivars, different climates and cropping systems. The distances among the five populations ranged from 175 to 1,007 km (Fig. 1, Table 1).
To confirm the results of the morphological evaluation, molecular identification was performed using the tef1-α gene and ITS-rDNA regions. Based on the obtained results, 50 selected isolates belonged to the M. phaseolina species (Supplementary Table 1). New nucleotide sequences described in this manuscript are available for download through GenBank under accession numbers OM362558 to OM362607 for the tef1-α gene and OM281348 to OM281397 for ITS-rDNA region (Supplementary Table 1).
Measures of genetic diversity.
Diversity parameters for the five Iranian populations are summarized in Table 2. Five out of seven ISSR primers were successfully amplified fragments with sizes rangingfrom 250 to 2,600 bp among M. phaseolina isolates in Iranian populations (Supplementary Table 2). All loci were polymorphic. The number of alleles ranges from 8 to 16 at each locus (Table 2). Gene diversity ranged from 0.58 (ISSR10) to 0.83 (ISSR02) with an average of 0.68 across all loci and was significantly different at P≤0.05 among loci (Table 2). The analysis of ISSR revealed high levels of gene and genotype diversity and allelic richness in each Iranian field population (Table 3). A total of 152 different multilocus genotypes were identified among the 200 Iranian isolates. The degree of genotypic diversity ranged from 43% (Kogiluyeh & Boyer-Ahmad) to 98% (Fars). The Nei's measure of gene diversity varied between 0.46 in Kogiluyeh & Boyer-Ahmad to 0.84 in Fars and averaged 0.68 across all populations (Table 3). Differences in allelic richness among M. phaseolina populations were calculated by resampling 100 data sets. The allelic richness ranged from 2.94 in the Kogiluyeh & Boyer-Ahmad population to 6.10 in the Fars population, and the average allelic richness across all populations was 4.80 (Table 3).
AMOVA and pairwise population differentiation.
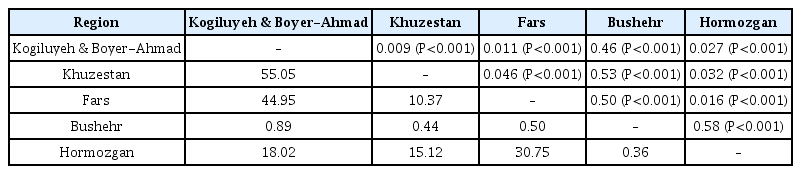
An AMOVA was used to explain the hierarchical distribution of Iranian M. phaseolina populations. Analyses of the distribution of gene diversity within and among five Iranian populations were 61% and 39%, respectively. Genetic differentiation was estimated between all Iranian populations with F-statistic using FST (Table 4). Genetic differentiation between all population pairs ranged from FST=0.009 to 0.58. The lowest and the highest FST were between the population pair Khuzestan-Kogiluyeh & Boyer-Ahmad (0.009) and Hormozgan-Bushehr (0.58), respectively (Table 4). While the effective migration rate (Nm) between all populations pair was inverse FST, Nm ranging from 0.36 to 55.05. The lowest and the highest Nm were between the population pair Hormozgan-Bushehr (0.36) and Khuzestan-Kogiluyeh & Boyer-Ahmad (55.05), respectively (Table 4).
Population structure analysis.
The STRUCTURE analysis clustered five Iranian populations into two distinct groups (Fig. 3). The assignment analysis revealed that the individuals from group 1 were assigned to the Bushehr population from which it was sampled. In contrast, the individuals from Khuzestan, Fars, Hormozgan, and Kohgiluyeh & Boyer-Ahmad were aggregated and formed group 2 even though different geographical origins, suggesting high gene flow between these four populations of group 2. Few isolates from group 1 were identified as being admixed mainly with group 2, suggesting low gene flow between these two groups.
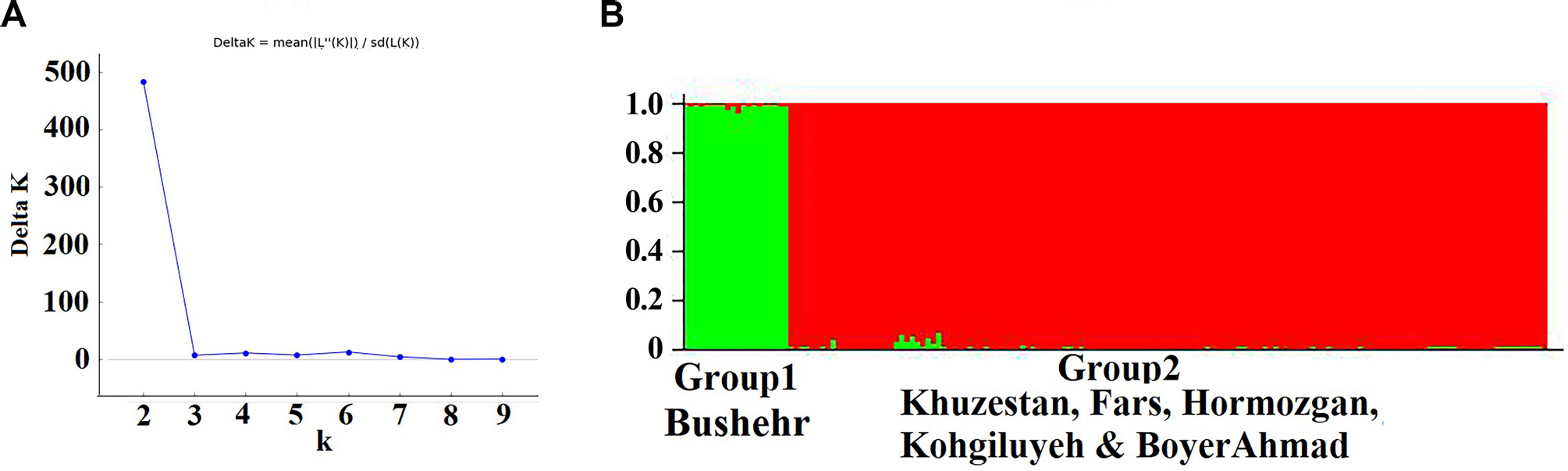
Structure analysis of the Iranian Macrophomina phaseolina populations. Individuals were assigned to clusters based on inter simple sequence repeats marker. (A) ΔK indicates the highest likelihood at K=2 supposed populations. (B) The STRUCTURE plot clustered the Iranian M. phaseolina populations from different locations into two distinct groups (K=2).
Discussion
Charcoal rot disease is an important fungal disease of sesame and represents a serious threat to sesame production in Iran because of climate change scenarios of drought stress and increased heat. The lack of information on the genetic structure of M. phaseolina populations has highlighted the need to perform this research. To the best of our knowledge, the present work is a study of population structure of a large collection of M. phaseolina isolates collected from five different climatic regions in Iran; temperate with mild winters (Fars and Kogiluyeh & Boyer-Ahmad provinces), warm and dry winter (Khuzestan province), and warm, dry with cool winter (Bushehr and Hormozgan provinces). ISSR marker has been extensively used for assessing variation within and among M. phaseolina species (Khamari et al., 2023; Khamari et al., 2022; Mahdizadeh et al., 2012; Salahlou et al., 2016; Yousef, 2021). In the present study, results have shown a high level of polymorphism among M. phaseolina populations of sesame with different geographical origins using neutral ISSR marker in southern Iran. These findings are consistent with previous studies (Aghakhani and Dubey, 2009; Khamari et al., 2023; Khamari et al., 2022; Linhai et al., 2011; Yousef, 2021) that have demonstrated high genetic diversity of M. phaseolina is reflected not only in isolates from different hosts but also within isolates collected from a special host or geographical origin. In general, high genetic diversity levels among a large collection of M. phaseolina populations could be attributed to para-sexual recombination between nuclear genes or fusion of vegetative cells favoring heterokaryons, this is corroboration with earlier studies (Carlile, 1986). In contrast, the low genetic diversity observed among M. phaseolina populations in Iran by Salahlou et al. (2016), is probably due to the small population (3 to 12 isolates) collected in each province, which is not a suitable measure to evaluate the genetic structure.
One of the important findings of this study was the detection of high genetic differentiation (FST: 0.46 to 0.58) and limited effective migration rate (Nm: 0.36 to 0.89) between the Bushehr population with four other populations that geographical distance was from 297 to 727 km. Therefore, the M. phaseolina population from Bushehr is genetically differentiated from the other populations and is the most genetically divergent of these four populations. In contrast, there was low genetic differentiation among four populations; Fars, Kogiluyeh & Boyer-Ahmad, Khuzestan and Hormozgan (FST: 0.009 to 0.046), even though the geographical distances range from 175 to 1,007 km. The overall migration rate (Nm) across the four populations was from 10.37 to 55.05, indicating high genetic exchange occurs between four populations and there are few barriers to genetic exchange (Table 4). However, geographical distance may always not play an important role in the observed differentiation (Stukenbrock et al., 2006). Population analysis using STRUCTURE revealed that Iranian isolates were subdivided into two populations (Fig. 3). The results matched with genetic differentiation and gene flow among four regions (Table 4).
We were able to isolate 10 M. phaseolina isolates from seeds of symptomatic sesame plants in this research. Since M. phaseolina could survive for up to 3 years as microsclerotia in symptomatic seeds or as mycelium in asymptomatic seeds (Ghosh et al., 2018; Londoño, 2022; Vashisht et al., 2023). Consequently, it is supposed that low genetic differentiation with high gene flow may be associated with the movement of infected seeds and soil with planting materials by vegetative stage and microsclerotia or airborne ascospores may provide an important source of primary infection (Ghosh et al., 2018; Salahlou et al., 2016). The sexual stage of M. phaseolina has only rarely been seen in nature (Ghosh et al., 2018), and it has been suggested that primary infection via infected seed coupled with human mediated dispersal of infected seed over long distances could be a major contributor to the observed patterns of genetic variability. Hence, infected seeds are thought to be the dominant mechanism responsible for the dispersal of the pathogen in southern Iran. Mostly, the scattering of plant pathogens over long distances can have significant consequences for disease management in agricultural ecosystems (Brown and Hovmøller, 2002). It should be noted that, seed exchange is rarely done between Bushehr province with other provinces, and farmers usually use seeds from previous years. In addition, there is a diversity of different plants that are cultivated in this province and may be host of M. phaseolina, which needs further investigation.
We expected minor genetic variation among Iranian M. phaseolina populations because of crop rotation that may diminish pathogen propagules and limit somatic recombination (Almeida et al., 2003; Rayatpanah et al., 2012). While, the present study reveals high genetic diversity among M. phaseolina populations of sesame in southern Iran. Generally, factors such as immigration, climatic conditions, high selection pressure, breeding system, and wide host range within the country have been used to clarify high levels of genetic diversity in the Iranian M. phaseolina populations. Our findings were mostly in agreement with these earlier studies (Almeida et al., 2003; Mahdizadeh et al., 2012; Purkayastha et al., 2006; Salahlou et al., 2016). Despite isolating only one M. phaseolina species of sesame and due to broad range of host plants and geographical distribution of this species from Iran (Ershad, 2009; Salahlou et al., 2016), suggests that this fungus is quite heterogeneous. This variability is observed in the majority of isolates obtained from the sesame plant. Consequently, genetic features of this pathogen are of importance for effective disease management versus a low variation observed in morphological characterizations.
The present work provided information about genetic structure in a large collection of M. phaseolina populations from five different geographical regions in Iran, and determined that ISSR marker is an appropriate molecular tool for assessing genetic diversity within and between M. phaseolina populations. Gene flow was mostly responsible for shaping or structuring M. phaseolina populations in Iran. If gene flow continues, it will be difficult to manage charcoal rot disease in different geographical regions of Iran. Our results are consistent with Almeida et al. (2023) who reported that the high genetic diversity among M. pseudophaseolina isolates makes it hard to improve resistant melon varieties to Macrophomina isolates. In summary, it is essential to have local quarantine and prevent seed exchanges between geographical populations to restrict the dispersal of pathogen over long distances and provide certified seeds in Iran.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
The authors thank the Yasouj University for financial support.
Electronic Supplementary Material
Supplementary materials are available at Research in Plant Disease website (http://www.online-rpd.org/).
Supplementary Table 1.
Macrophomina isolates used in order to molecular identification
Supplementary Table 2.
Primer names and sequences, annealing temperature of amplified DNA using each of the 5 primers of ISSR in this study