In 2020, the production area of cucumber (
Cucumis sativus) was 4,721 ha with an annual yield of 335,596 tons in Korea (
Korean Statistical Information Service, 2022). However, cucumbers are susceptible to various diseases during their growth period, including soft rot caused by
Pectobacterium species, which can result in significant economic losses for agricultural production. Several species of
Pectobacterium, including
P. brasiliense and
P. carotovorum subsp.
carotovorum, have been reported as causative agents of cucumber soft rot in China and Malaysia (
Meng et al., 2017;
Nazerian et al., 2011). Soft rot caused by various strains of
P. brasiliense has been reported in Asian countries, including China, Japan, and Korea, affecting a variety of plants, such as cucumber, graft-cactus, napa cabbage, potato, radish, and tobacco (
Fujimoto et al., 2017;
Jee et al., 2020;
Meng et al., 2017;
Park et al., 2022,
2023;
Wang et al., 2017).
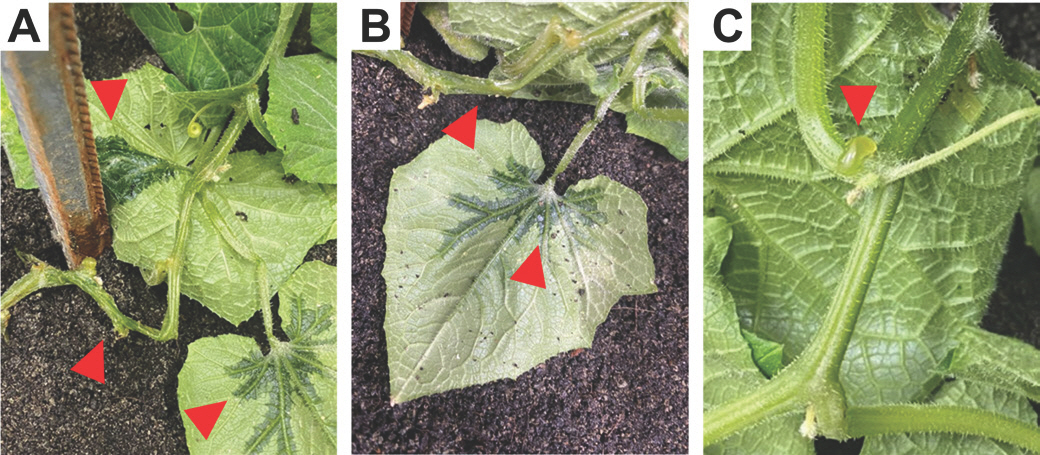
In June 2021, numerous cucumber samples exhibiting wilted and water-soaked lesions symptoms were collected from Daejeon, Chungcheongnam-do, Korea. The affected cucumber plants showed symptoms of soft rot on the surface of infected tissues, water soaking in infected stems, and wet rot on the edges or centers of leaves (
Fig. 1A,
B). Furthermore, conspicuous symptoms of liquid production became apparent on the infected stem, indicating the presence of exudates or secretions resulting from the infection (
Fig. 1C). These symptoms, reminiscent of bacterial soft rot as described by
Charkowski (2018), provide visible evidence of the pathogenic impact on the stem tissue. The presence of liquid production on the infected stems serves as a characteristic hallmark, reinforcing the resemblance to bacterial soft rot and confirming the manifestation of the disease symptoms.
Fig. 1.
Symptoms of soft rot disease caused by Pectobacterium brasiliense on cucumber plants in Daejeon, Chungcheongnam-do, Korea. (A, B) Stems and leaves of cucumber were wilted and have water-soaked lesions caused by P. brasiliense KNUB-04-21. (C) Liq-uid production was observed from the infected stem. Arrowheads indicate the invasion of soft rot.
To isolate the pathogenic bacterial agent, cucumber tissues exhibiting symptoms of infection were immersed in a 5 ml saline solution (0.85% NaCl) for 20 min. Subsequently, a 50 μl aliquot of the resulting suspension was uniformly spread onto nutrient agar (NA; Difco, Frankin Lakes, NJ, USA) medium and incubated for 24 hr at 27 ° C. After the purification process using NA and streaking on King's medium B (Difco), the bacterial strain was successfully isolated from samples collected in the region of Daejeon and was designated KNUB-04-21, serving for further analyses.
For molecular analysis, the total genomic DNA of KNUB-04-21 was extracted from 24 hr cultures grown on NA using HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea). The polymerase chain reaction (PCR) protocol used in this study for amplifying the 16S rRNA region was based on the methodology described by
Weisburg et al. (1991). The 16S rRNA region of KNUB-04-21 (GenBank no. LC744204) was 1,352 bp in its sequence length. After a BLAST search in the National Center for Biotechnology Information (NCBI) database, it was found that the 16S rRNA gene sequence of KNUB-04-21 displayed similarities of 99.78% and 99.70% with that of
P. brasiliense BP201611 (GenBank no. MN393922) and
P. carotovorum subsp.
carotovorum PDP201711 (GenBank no. MN394009), respectively. These findings clearly demonstrated that relying solely on the comparative analysis of the 16S rRNA gene sequence was insufficient to accurately identify the strain KNUB-04-21 at the species level. These data also underscore the limitations of using the 16S rRNA gene as a standalone marker for species identification and emphasize the need for using additional molecular markers or comprehensive genomic approaches to achieve a more precise and reliable taxonomic classification. Therefore, following
Portier et al. (2019), the genes
dnaX, leuS, and
recA of KNUB-04-21 were subjected to PCR amplification and sequencing.
The PCR protocols used in this study to amplify
dnaX, leuS, and
recA were based on the methodology outlined by
Sławiak et al. (2009),
Portier et al. (2019), and
Waleron et al. (2002), respectively (
Supplementary Table 1). Fragments con-taining 509, 518, and 715 bp were obtained for
dnaX, leuS, and
recA, respectively. All obtained gene sequences were registered in the NCBI with the following GenBank accession numbers:
dnaX, LC744201;
leuS, LC744202;
recA, LC744203. Based on the similarity of the
dnaX sequence, KNUB-04-21 exhibited a similarity of >98% with
P. brasiliense NY1563A (GenBank no. MW979072) and
P. carotovorum subsp.
carotovorum PCC21 (GenBank no. CP003776). The
leuS sequence also exhibited a similarity of >98% with
P. brasiliense 130 (GenBank no. CP092039),
P. carotovorum subsp.
carotovorum PCC21 (GenBank no. CP003776), and
P. quasiaquaticum A398-S21-F17 (GenBank no. CP065178). Furthermore, the
recA sequence of KNUB-04-21 demonstrated a similarity of >98% with
P. brasiliense CFBP6615 (GenBank no. MK517243) and
P. carotovorum subsp.
carotovorum 333 (GenBank no. AY264787).
The NCBI BLAST was used to compare the sequences of the three housekeeping genes with those of the reference
Pectobacterium species in the genomic databases (
Table 1). Phylogenetic and molecular evolutionary analyses were conducted using the MEGA 7 software (
Kumar et al., 2016). Maximum likelihood analysis was performed using Kimura's two-parameter model and the nearest neighbor interchange heuristic search method. The reliability and robustness of the obtained clusters were evaluated using bootstrap analysis with 1,000 replicates. The constructed phylogenetic tree revealed that KNUB-04-21 and multiple
P. brasiliense strains formed a monophyletic clade with a high bootstrap value, strongly suggesting that they belong to the same species (
Fig. 2).
Fig. 2.
Maximum-likelihood phylogenetic tree, based on concatenated partial sequences of dnaX, leuS, and recA genes, showing the phylogenetic position of Pectobacterium brasiliense KNUB-04-21 among related species of the genus Pectobacterium. Bootstrap values (based on 1,000 replications) greater than 70% are shown at branch points. Dickeya solani CFBP7704 was used as the outgroup. The scale bar repre-sents 0.020 substitutions per nucleotide position.
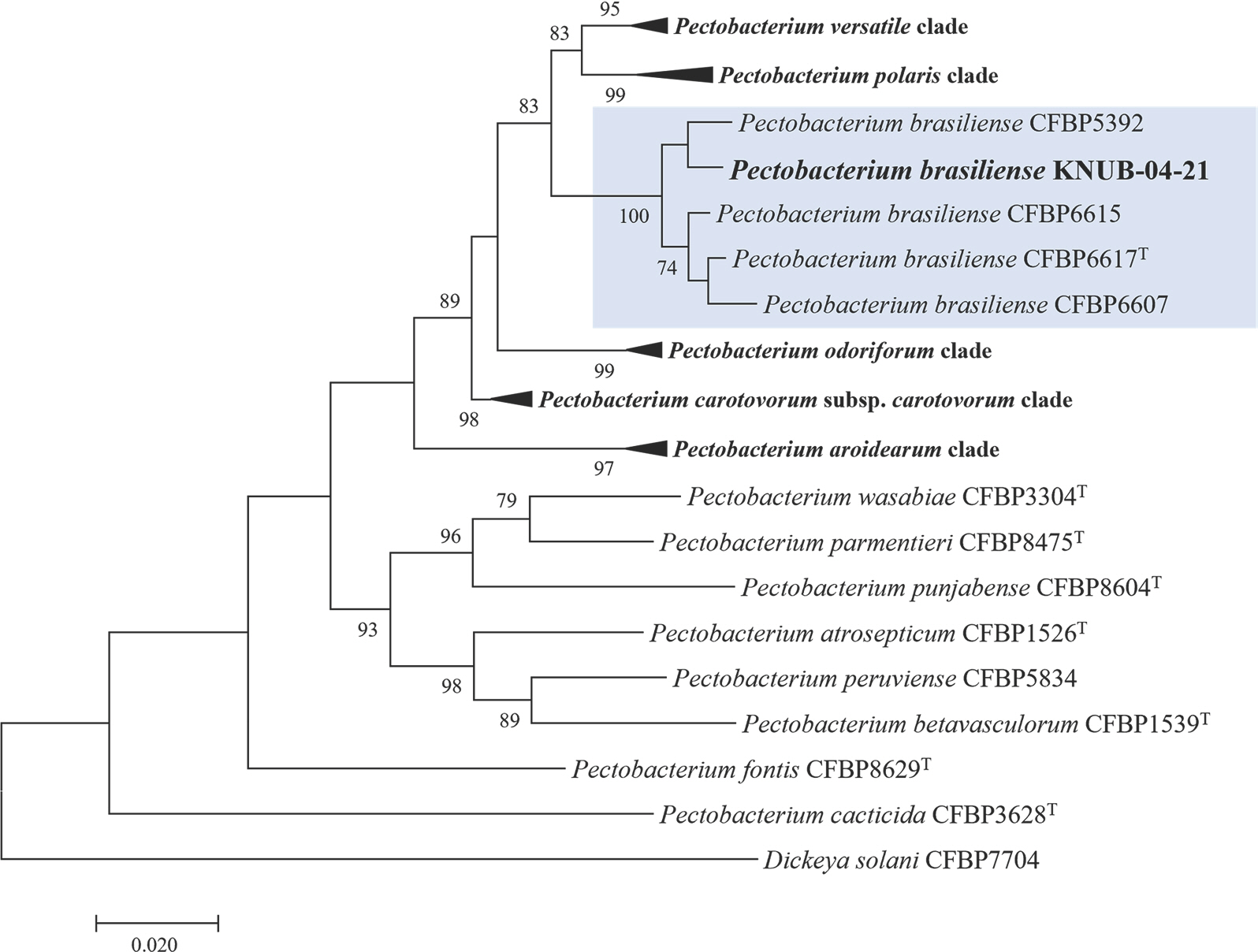
Table 1.
List of Pectobacterium species and Dickeya solani used in phylogenetic analysis with their corresponding GenBank accession numbers
|
Species |
Strain no. |
GenBank accession no. |
|
dnaX
|
leuS
|
recA
|
|
Pectobacterium aroidearum
|
CFBP1457 |
MT683925 |
MT684072 |
MT684219 |
|
Pectobacterium aroidearum
|
CFBP2573 |
MT683941 |
MT684088 |
MT684235 |
|
Pectobacterium aroidearum
|
CFBP6725 |
MT684029 |
MT684176 |
MT684323 |
|
Pectobacterium aroidearum
|
CFBP8737 |
MT684054 |
MT684201 |
MT684348 |
|
Pectobacterium atrosepticum
|
CFBP1526 T
|
MK516904 |
MK517048 |
MK517192 |
|
Pectobacterium betavasculorum
|
CFBP1539 T
|
MK516905 |
MK517049 |
MK517193 |
|
Pectobacterium cacticida
|
CFBP3628 T
|
MK516923 |
MK517067 |
MK517211 |
|
Pectobacterium brasiliense
|
KNUB-04-21
|
LC744201
|
LC744202
|
LC744203
|
|
Pectobacterium brasiliense
|
CFBP5392 |
MK516927 |
MK517071 |
MK517215 |
|
Pectobacterium brasiliense
|
CFBP6607 |
MK516954 |
MK517098 |
MK517242 |
|
Pectobacterium brasiliense
|
CFBP6615 |
MK516955 |
MK517099 |
MK517243 |
|
Pectobacterium brasiliense
|
CFBP6617 T
|
MK516956 |
MK517100 |
MK517244 |
|
Pectobacterium carotovorum subsp. carotovorum
|
CFBP1364 |
MK516896 |
MK517040 |
MK517184 |
|
Pectobacterium carotovorum subsp. carotovorum
|
CFBP2046 T
|
MK516909 |
MK517053 |
MK517197 |
|
Pectobacterium carotovorum subsp. carotovorum
|
CFBP6071 |
MK516950 |
MK517094 |
MK517238 |
|
Pectobacterium carotovorum subsp. carotovorum
|
CFBP7351 |
MK516962 |
MK517106 |
MK517250 |
|
Pectobacterium odoriferum
|
CFBP1878 T
|
MK516907 |
MK517051 |
MK517195 |
|
Pectobacterium odoriferum
|
CFBP3259 |
MK516920 |
MK517064 |
MK517208 |
|
Pectobacterium odoriferum
|
CFBP3297 |
MK516921 |
MK517065 |
MK517209 |
|
Pectobacterium odoriferum
|
CFBP5539 |
MK516929 |
MK517073 |
MK517217 |
|
Pectobacterium fontis
|
CFBP8629 T
|
MK516878 |
MK517022 |
MK517166 |
|
Pectobacterium parmentieri
|
CFBP8475 T
|
MK516972 |
MK517116 |
MK517260 |
|
Pectobacterium peruviense
|
CFBP5834 |
MK516935 |
MK517079 |
MK517223 |
|
Pectobacterium polaris
|
CFBP1403 |
MK516898 |
MK517042 |
MK517186 |
|
Pectobacterium polaris
|
CFBP6058 |
MK516945 |
MK517089 |
MK517233 |
|
Pectobacterium polaris
|
CFBP7360 |
MT684038 |
MT684185 |
MT684332 |
|
Pectobacterium polaris
|
CFBP8603 T
|
MT684046 |
MT684193 |
MT684340 |
|
Pectobacterium punjabense
|
CFBP8604 T
|
MK516877 |
MK517021 |
MK517165 |
|
Pectobacterium versatile
|
CFBP1118 |
MK516888 |
MK517032 |
MK517176 |
|
Pectobacterium versatile
|
CFBP2138 |
MK516912 |
MK517056 |
MK517200 |
|
Pectobacterium versatile
|
CFBP6051 T
|
MK516938 |
MK517082 |
MK517226 |
|
Pectobacterium versatile
|
CFBP8656 |
MK516973 |
MK517117 |
MK517261 |
|
Pectobacterium wasabiae
|
CFBP3304 T
|
MK516922 |
MK517066 |
MK517210 |
|
Dickeya solani
|
CFBP7704 |
MK516970 |
MK517114 |
MK517258 |
To characterize the strain KNUB-04-21, it was subjected to a comprehensive analysis using the API ID 32 GN system (Biomérieux, Marcy l'Etoile, France) according to standard-ized procedures recommended by the manufacturer. The inoculum of KNUB-04-21 was prepared by selecting a colony from the surface of NA medium. KNUB-04-21 demonstrated positive assimilation responses to various compounds, including D-glucose, D-maltose, D-mannitol, D-melibiose, inositol, L-alanine, L-rhamnose, L-serine, N-acetylglucosamine, salicin, and sucrose. Conversely, KNUB-04-21 exhibited negative utilization responses to 3-hydroxybutyric acid, D-sorbitol, L-fucose, L-histidine, and propionic acid. These findings are consistent with the characteristics outlined by
Portier et al. (2019) for the reference strain
P. brasiliense.
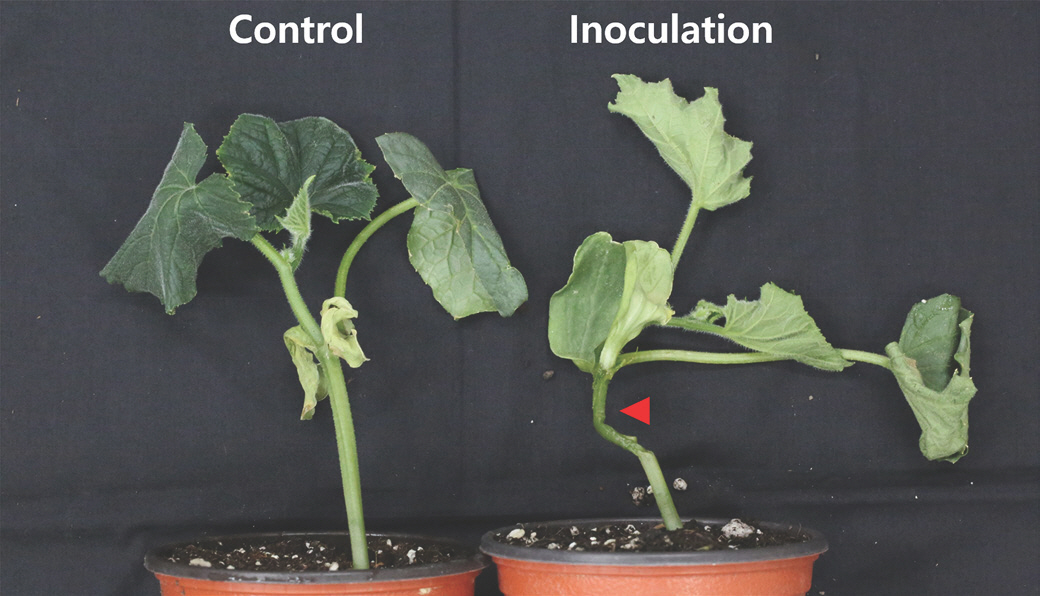
To fulfill Koch's postulates and establish the pathogenicity of KNUB-04-21, pathogenicity tests were conducted on cucumber plants. Before inoculation, the cucumber surface was sterilized using 70% ethanol, followed by rinsing with distilled water. A 1 ml suspension of KNUB-04-21 with a concentration of 1×10
8 cells/ml was used for inoculation. As a control, a mock infection was performed by inoculating the cucumber with 1 ml of distilled water. The experiment was conducted under controlled greenhouse conditions, maintaining a temperature range of 25-30 ° C and a relative humidity of 80% throughout the study period. At 2 days after inoculation, the cucumber stems exhibited characteristic symptoms of wilting and water-soaked lesions, as shown in
Fig. 3. In contrast, the cucumber stems subjected to mock infection exhibited no symptoms (
Fig. 3). Reidentification of the bacterial strain isolated from each symptomatic cucumber confirmed its classification as
P. brasiliense, although the details of this reidentification are not described in this report. These data provide strong evidence supporting the phyto-pathogenic nature of KNUB-04-21 and its association with the observed disease symptoms in the cucumber plants.
Fig. 3.
Effect of inoculating Pectobacterium brasiliense KNUB-04-21 on the cucumber stem. Two days after inoculation, symptoms such as wilting and water-soaked lesions were observed in the stem of the cucumber. However, no symptoms were observed in the control plant. Arrowheads indicate the invasion of bacterial cells and soft rot areas.
Several species of
Pectobacterium have been identified as disease-causing agents for various crops. Among them,
P. brasiliense has been reported to infect 19 different plant species belonging to 10 families (
Oulghazi et al., 2021). In Korea, the first detection of
P. brasiliense was performed in 2012 (
Choi and Kim, 2013). Furthermore,
P. brasiliense is a pathogen of amaranth, Chinese cabbage, chrysanthemums, eggplant, graft-cactus, napa cabbage, nepenthes, paprika, pepper, potato, radish, and tomato (
Choi and Kim, 2013;
Jee et al., 2018,
2020;
Lee et al., 2014;
Park et al., 2022,
2023). Low-cost, sensitive, and specific methods were recently devel-oped for efficient detection of target pathogens belonging to the genus
Pectobacterium (
Jin et al., 2022;
Oulghazi et al., 2021). This study describes the identification of
P. brasiliense as the causal agent of cucumber soft rot in Korea. This finding not only expands our understanding of the distribution of
P. brasiliense in Korea but also enhances our knowledge of the range of causative agents responsible for cucumber soft rot. Moreover, this study can contribute to the development of more efficient management strategies for minimizing the economic damage caused by this disease.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print